Abstract
In this study, the mitogenomes of four smelts (Hypomesus nipponensis) were sequenced and the genomic variation and phylogeny were examined. The mitogenome consisted of 16,782 bp and showed 99.5 ∼ 99.7% sequence similarity with the previously released sequence. A total of 130 single nucleotide polymorphisms were detected and they were present in 2 rRNA genes, 6 tRNA genes, 12 protein-coding genes, and 1 non-coding region. This study provides important information for the classification of H. nipponensis and related species.
The smelt, Hypomesus nipponensis, is a small fish species in the order, Osmeriformes within the family, Osmeridae. It is distributed in the North Pacific region, including in Japan and the Korean peninsula (Katayama et al. Citation2001; Ilves and Taylor Citation2008). This fish is favored by Koreans as winter food. Since 1997, some regions have held annual smelt-themed ice-fishing festivals, creating hundreds of thousands of dollars of direct economic impact. Previously, the H. nipponensis genome was released to the NCBI database under GenBank accession number, HM106489.1, but its details were not published as a manuscript. We assembled four smelt mitochondrial genomes with the released sequence as a reference. In the present study, we sequenced the mitochondrial DNA of four individuals and examined variations in their mitogenome sequence when compared with the reference sequence (HM106489.1).
Four smelts were obtained from Lake Daechung (N 36.491367° E 127.491606°) in the Republic of Korea. Genomic DNA from four muscle tissue samples was extracted using a DNeasy Tissue Kit (Qiagen, Valencia, CA, USA). DNA libraries were constructed using a Nextera XT sample preparation kit (Illumina, San Diego, CA, USA) and sequenced on an Illumina HiSeq 2000 platform by Macrogen (Seoul, Republic of Korea). Final assembled sequences were further annotated using the web-based automatic annotation server, MitoFish (Iwasaki et al. Citation2013) and single nucleotide polymorphisms (SNPs) were detected with the DnaSP program (Librado and Rozas Citation2009). Non-synonymous SNPs were manually searched by translating protein-coding genes (PCGs) using MEGA7 (Kumar et al. Citation2016). The complete sequence, with annotations, has been deposited in GenBank under the accession numbers MK234844–47. A phylogenetic tree was constructed with the Cox1 gene of 4 H. nipponensis samples, including previously reported sequences and 10 species belonging to the order, Osmeriformes.
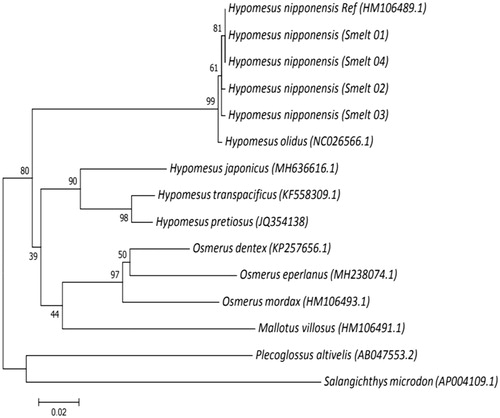
The H. nipponensis mitochondrial genome was 16,782 bp in length, had 37 genes (2 rRNA genes, 22 tRNA genes, and 13 PCGs), and control regions that are typically observed in fish mitochondrial genomes (Li et al. Citation2010). Nine genes (ND6, tRNA-Gln, Ala, Asn, Cys, Tyr, Ser, Glu, and Pro) were encoded on the light strand and the others were encoded on the heavy strand, as observed in H. nipponensis. The nucleotide composition of the mitochondrial genome was 24.35% A, 27.33% T, 19.18% G, and 29.12% C. The observed 48.30% G + C content level was similar to other smelts (e.g. Hypomesus olidus, NC_026566.1, 48.34%; Hypomesus joponicus, MH636616.1, 48.7%; and Osemerus eperlanus, MH238074.1, 47.56%). All PCGs, except Cox1, were initiated by the common ATG start codon, while the start codon for Cox1 was GTG. Five of the PCGs (ND1, Cox1, ATP8, ND4L, and ND5) were terminated by the stop codon, TAA. In total, 130 SNPs were detected (0.775% of the mitogenome sequence) and they were located in both rRNAs (12S and 16S rRNA), 5 tRNAs (Tyr, Ser, Glu, Thr, and Pro), and 12 PCGs (Cox1–3, ND1–6, ATP8, ATP6, and Cyt b). There were no SNPs detected in ND4L. Twelve SNPs in six of the polymorphic PCGs (ND2–6 and ATP6) were considered to be non-synonymous SNPs. Phylogenetic analysis using the Cox1 gene confirmed the current taxonomic classification of H. nipponensis (). In fact, the mitogenome of H. nipponensis exihibited 99% sequence similarity with H. olidus, with the same gene arrangement, composition, and size (Li et al. Citation2010; Bai et al. Citation2017). As can be seen in , the Cox1 gene is also very similar between the two species. The data presented here will be an important resource for the identification of H. olidus and H. nipponensis species in subsequent studies. The smelt genomic DNA used in this study is deposited in National Marine Biodiversity Institute of Korea under the Ministry of Oceans and Fisheries (sample No. MABIK GR00002108).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bai X, Luo Z, Feng S, Sun Z, Wu H, Li C, Jiang J, Liu X, Wang N. 2017. The complete mitochondrial genome of Hypomesus olidus (Osmeriformes: Osmeridae). Mitochondr DNA. 2:687–688.
- Ilves KL, Taylor EB. 2008. Evolutionary and biogeographical patterns within the smelt genus Hypomesus in the North Pacific Ocean. J Biogeogr. 35:48–64.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: A mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Katayama S, Kijima A, OMoRI M, Okata A. 2001. Genetic differentiation among and within local regions in the pond smelt. Hypomesus nipponensis. Tohoku J Agric Res. 51:61–77.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Li J, Xia R, McDowall R, López JA, Lei G, Fu C. 2010. Phylogenetic position of the enigmatic Lepidogalaxias salamandroides with comment on the orders of lower euteleostean fishes. Mol Phylogenet Evol. 57:932–936.
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452.