Abstract
In this study, we determined the complete mitogenome of Sphaeroma terebrans (Crustacea, Isopod, Sphaeromatidae), which was the first one in the family Sphaeromatidae. The mitogenome of S. terebrans was 15,540 bp in length and contained 13 protein-coding genes, 21 tRNA genes, 2 rRNA genes, 1 control region, and 2 unknown fragments which is longer than 200 bp. The overall base composition was 30.1% A, 14.9% C, 21.3% G, and 33.7% T. Six start codons (ATA, ATC, ATG, ATT, ACG, GTG) and three stop codons (TAA, TAG, T–) were found in the protein-coding genes. The size of 21 tRNA genes ranged from 51 to 66 bp, tRNA-Gly was not found. All nodes strongly supported that S. terebrans was placed as sister to Sphaeroma serratum in the maximum likelihood tree.
Sphaeroma terebrans (Crustacea, Isopod, Sphaeromatidae) is widely distributed in tropics and subtropics areas (Harrison and Holdich Citation1984; Villalobos et al. Citation1985). It is a small marine wood-boring isopod, usually colonises in the roots and aerial roots of mangrove, sedimentary rocks, foam and other materials, which mainly feeds on plankton (Si et al. Citation2002). The species is also a pest that can destroy mangrove plants (Svavarsson et al. Citation2002). In this study, we first determined the complete mitogenome of S. terebrans and analyzed the phylogenetic position of this species.
A group samples of S. terebrans were collected from one mangrove hole in Lianzhou Bay (21°33′N, 109°09′E), Beihai, Guangxi Province, China, and preserved in 95% ethanol. One specimen was selected for DNA extraction and deposited in Guangxi Mangrove Research Center with a voucher number BH20160701 and the others were preserved for morphological identification. The total DNA was isolated using E. Z. N. A.TMSQ Tissue DNA Kit (Omega, Norcross, Georgia, USA), and sequenced by Illumina high-throughput sequencing. The protein-coding genes of S. terebrans were annotated with Dogma (Wyman et al. Citation2004) and boundaries of protein-coding genes and rRNA genes were determined by sequence comparison with other Isopod species. All tRNA genes were identified using tRNAscan-SE1.2.1 (Lowe and Chan Citation2016), Dogma (Wyman et al. Citation2004), and ARWEN (Laslett and Canbäck Citation2008) in the default search mode, and folded using RNAstructure 5.2 (Reuter and Mathews Citation2010). Including S. terebrans, 11 species of Isopod with mitogenomes available in GenBank were used for the phylogenetic reconstruction. Metacrangonyx longipes was selected as an outgroup. Phylogenetic analyses were conducted using the maximum likelihood (ML) by PHYML 3.0 (Guindon et al. Citation2010) with 1000 bootstrap replicates under the MtArt model. All analyses were performed with the concatenated amino acid sequences of the 13 protein-coding genes.
The complete mitogenome of S. terebrans was 15, 540 bp in length (GenBank MK460228). The overall base composition was 30.1% A, 14.9% C, 21.3% G, and 33.7% T. It consisted of 13 protein-coding genes, 21 transfer RNA genes, 2 rRNA genes, 1 control region, and 2 unknown fragments which is longer than 200 bp. In S. terebrans, the protein-coding genes, COX1-3, ATP6, ATP8, ND2, ND3, ND5, ND6 were encoded on the H-strand, while CYTB, ND1, ND4, ND4L were encoded on the L-strand. This arrangement of protein-coding genes was in the same way as Ligia oceanica (Kilpert and Podsiadlowski Citation2010). All the protein-coding genes started with ATN codons except for ND3 and COX1, which used GTG and ACG, respectively. Although using ACG as the initiation codon seemed to be unusual in metazoan mitogenome, it was another prevalent start codon for the COX1 gene in malacostracan crustaceans (Kilpert and Podsiadlowski Citation2006, Citation2010). All the protein-coding genes used TAA, TAG, and T– as termination codons. The 12S rRNA and 16S rRNA genes are 739 bp and 1227 bp in length, respectively. The size of 21 tRNA genes ranged from 51 to 66 bp. Although we did a lot of work to search secondary structures in non-coding regions and control region, the tRNA-Gly gene was not found in the mitogenome. All of the tRNA genes could fold into a typical clover-leaf secondary structure except for tRNA-Leu (UAG), tRNA-Cys, and tRNA-Ile genes replacing the dihydrouridine arm by a simple loop. The control region (742 bp) was located between the tRNA-Gln and tRNA-Glu genes, rich in A + T content (68.6%).
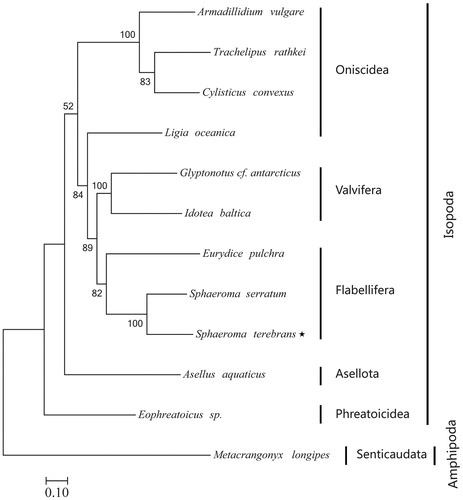
In the maximum likelihood tree, most nodes were well supported (). Eleven available species belonged to five suborder with the topology (Phreatoicidea (Asellota (Oniscidea (Valvifera + Flabellifera)))), and the suborder Phreatoicidea was the basal clade. Sphaeroma terebrans was clustered to S. serratum with high support value (100%), then this clade clustered to Eurydice pulchra, and all of them belonged to suborder Flabellifera. This result was consistent with morphological taxonomy (Liu Citation2008; Yu and Li Citation2003). In addition, Ligia oceanica, which was classed to the suborder Oniscidea in morphological view, clustered to the species of the suborder Valvifera and Flabellifera. Therefore, the relationship within the order Isopoda needs further study with more species.
Figure 1. Phylogenetic position of Sphaeroma terebrans (Asterisked). Metacrangonyx longipes (AM944817) was selected as the out group. The 11 species from the order Isopod were S. terebrans (No. MK460228, asterisked), S. serratum (GU130256), Eurydice pulchra (GU130253), Glyptonotus cf. antarcticus (GU130254), Idotea baltica (DQ442915), Ligia oceanica (NC_008412), Cylisticus convexus (KR013002), Trachelipus rathkei (KR013001), Armadillidium vulgare (GU130251), Asellus aquaticus (GU130252), Eophreatoicus sp. (FJ790313).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Harrison K, Holdich DM. 1984. Hemibranchiate sphaeromatids (Crustacea: Isopoda) from Queensland, Australia, with a world-wide review of the genera discussed. Zool J Linn Soc. 81:275–387.
- Kilpert F, Podsiadlowski L. 2006. The complete mitochondrial genome of the common sea slater, Ligia oceanica (Crustacea, Isopoda) bears a novel gene order and unusual control region features. Bmc Genomics. 7:241.
- Kilpert F, Podsiadlowski L. 2010. The Australian fresh water isopod (Phreatoicidea: Isopoda) allows insights into the early mitogenomic evolution of isopods. Comp Biochem Physiol Part D Genomics Proteomics. 5:36–44.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Liu R. 2008. Checklist of marine biota of China seas. Beijing: Science Press; p. 772–809.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Reuter JS, Mathews DH. 2010. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 11:129.
- Si A, Bellwood O, Alexander CG. 2002. Evidence for filter-feeding by the wood-boring isopod, Sphaeroma terebrans (Crustacea: Peracarida). J Zool. 256:463–471.
- Svavarsson J, Osore MKW, Ólafsson E. 2002. Does the wood-borer Sphaeroma terebrans (Crustacea) shape the distribution of the mangrove Rhizophora mucronata? Ambio. 31:574–579.
- Villalobos CR, Cruz GA, Cruz RA. 1985. Notes on the biology of Sphaeroma terebrans Bate, 1866 (Sphaeromatidae: Isopoda) in the mangrove swamps of Pochote, Puntarenas Province, Costa Rica). Brenesia San Jose. 24:287–296.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yu HY, Li XZ. 2003. Study on the species of Sphaeromatidae from Chinese waters. Studia Marina Sinica. 45:239–259.
