Abstract
The complete mitochondrial genome was obtained by conventional polymerase chain reaction (PCR) from the intertidal hermit crab, Pagurus similis. The complete mitochondrial genome of P. similis was 15,683 bp in length, with the base composition of 36% A, 16% C, 11% G, and 37% T with a high AT bias of 73%. The assembled mitogenome contained 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and an AT-rich region. The gene order and contents were similar with previously reported Pagurus mitochondrial genomes. The complete P. similis mitogenome will provide an essential molecular reference to elucidate biogeography, phylogenetic distance, and evolutionary history in the genus Pagurus.
The family Paguridae (Decapoda: Anomura) exhibits high species diversity and are distributed worldwide in intertidal zones from tropical to temperate coastal waters. Since high-level taxonomic characterization of the genus Pagurus is complex due to morphologically heterogeneous characteristics and uncertain registration of many new species to this genus (Olguín and Mantelatto Citation2013), accumulation of the genomic information of Paguridae species is strongly needed to clarify their molecular phylogeny and genetic diversity (Bracken-Grissom et al. Citation2013). Despite the high species diversity and abundance of the genus Pagurus, only a few reports have been suggested on genomic characteristics and organization of whole mitogenomes in Paguridae species and studies on in-depth molecular phylogenetic relationship have yet to be conducted (Hickerson and Cunningham Citation2000; Sultana et al. Citation2018; Gong et al. Citation2019). Of genus Pagurus, whole mitochondrial genome information was firstly applied on P. longicarpus to understand genetic variation and unique gene rearrangements (Hickerson and Cunningham Citation2000). A wide range of genetic diversity of the genus Pagurus and their phylogenetic relationships was suggested using nearly complete mitochondrial genome sequences for 10 Pagurus species (Sultana et al. Citation2018). Recently, large-scale gene rearrangements were reported in the P. nigrofascia mitogenome and the phylogenetic relationship was suggested in the Anomura (Gong et al. Citation2019). In this study, the complete mitogenome of the intertidal hermit crab, P. similis (Ortmann, 1892) was analyzed as the first report. Although the morphometric analysis on P. similis was previously conducted (Komai Citation2003), there is no genomic information available on the species and study on the molecular phylogenetic evolution has not been extended as yet. Thus, more information on the complete mitochondrial genome is strongly needed for understanding the molecular evolution of Paguridae.
In this study, we sequenced the complete mitogenome of the intertidal hermit crab, P. similis (Accession no. MK673512). A single specimen of P. similis was isolated from the coastal region of Yeonpyeongdo (37°39′57.3″N, 125°40′49.9″E) and was preserved in 100% ethanol. The voucher specimen was deposited in the Marine Arthropod Depository Bank of Korea (MADBK; Species ID: 160724; Specimen ID: 160724_007). Genomic DNA was extracted from muscle tissue by using the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany). The genomic DNA was quantified using a Qubit 4 Fluorometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The species identity was verified by morphological assessment (Komai Citation2003) and sequence analysis of mitochondrial DNA cytochrome oxidase 1 (CO1) using a universal primer set (forward primer: 5′ ggt caa caa atc ata aag ata ttg g 3′; reverse primer: 5′ taa act tca ggg tga cca aaa aat ca 3′; Folmer et al. Citation1994). Whole mitogenome of P. similis was sequenced by Sanger-sequencing with specific primers based on the CO1 sequence. Overall sequences were annotated by using the MITOS web-based software (Bernt et al. Citation2013) and detailed annotation was conducted with NCBI-BLAST (http://blast.ncbi.nlm.nih.gov).
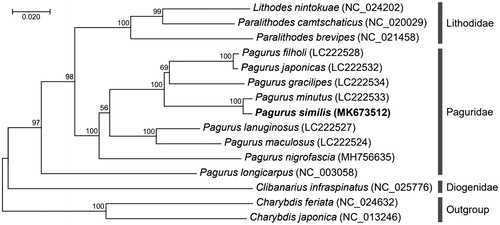
The complete mitogenome of P. similis was 15,683 bp in length and contained the typical set of 13 PCGs, 22 tRNAs, 2 rRNAs, and an AT-rich region. Overall, gene order and content of P. similis mitogenome were identical to those of Paguridae species (Hickerson and Cunningham Citation2000; Sultana et al. Citation2018; Gong et al. Citation2019). The nucleotide composition of P. similis mitogenome is heavily biased toward A + T nucleotides, accounting for 36% A, 16% C, 11% G, and 37% T. In particular, the highly A + T biased value (71.4%) was slightly higher than the value of P. nigrofascia mitogenome (Gong et al. Citation2019), as mainly differs in the AT-rich region. The AT-skew and GC-skew of P. similis mitogenome were –0.003 and –0.196, respectively, which were similar to those values identified in P. nigrofascia mitogenome (Gong et al. Citation2019). Phylogenetic tree was constructed on the concatenated set of nucleotide sequences of the 13 PCGs from the 9 mitochondrial genomes of Paguridae species (). In addition, three PCGs from family Lithodidae and one PCG from family Diogenidae were incorporated in phylogenetic analysis. By partitioned maximum likelihood analysis, the P. similis mitogenome was recovered as a well-supported clade [bootstrap percentage (BP) = 100] and clustered into a clade with other Paguridae species with high support value, which is consistent with the traditional taxonomy. In conclusion, the complete P. similis mitogenome will provide essential and fundamental information to elucidate phylogenetic distance, geographical distribution, and evolution of the genus Pagurus and related subspecies.
Figure 1. Maximum-likelihood (ML) phylogeny of 10 Paguridae species based on the concatenated nucleotide sequences of protein-coding genes (PCGs) with families Lithodidae and Diogenidae. Two Charybdis mitogenomes were used as AN outgroup for tree rooting based on the criteria of a previous publication (Gong et al. 2009). ML analysis was performed in RA + ML 8.2.10 using the mtREVþG model (Stamatakis Citation2014). Numbers on the branches indicate ML bootstrap percentages (1000 replicates). DDBJ/EMBL/Genbank accession numbers for published sequences are incorporated.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bernt A, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Bracken-Grissom HD, Cannon ME, Cabezas P, Feldmann RM, Schweitzer CE, Ahyong ST, Felder DL, Lemaitre R, Crandall KA. 2013. A comprehensive and integrative reconstruction of evolutionary history for Anomura (Crustacea: Decapoda). BMC Evol Biol. 13:128.
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Marine Biol Biotechnol. 3:294–299.
- Gong L, Jiang H, Zhu K, Lu X, Liu L, Liu B, Jiang L, Ye Y, Lü Z. 2019. Large-scale mitochondrial gene rearrangements in the hermit crab Pagurus nigrofascia and phylogenetic analysis of the Anomura. Gene. 695:75–83.
- Hickerson M, Cunningham C. 2000. Dramatic mitochondrial gene rearrangements in the hermit crab Pagurus longicarpus (Crustacea, Anomura). Mol Biol Evol. 17:639–644.
- Komai T. 2003. Identities of Pagurus japonicus (Stimpson, 1858) and P. similis (Ortmann, 1892), with description of a new species of Pagurus. Zoosystema. 25:377–411.
- Olguín N, Mantelatto FL. 2013. Molecular analysis validates of some informal morphological groups of Pagurus (Fabricius, 1775) (Anomura: Paguridae) from South America. Zootaxa. 3666:436–448.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Sultana Z, Asakura A, Kinjo S, Nozawa M, Nakano T, Ikeo K. 2018. Molecular phylogeny of ten intertidal hermit crabs of the genus Pagurus inferred from multiple mitochondrial genes, with special emphasis on the evolutionary relationship of Pagurus lanuginosus and Pagurus maculosus. Genetica. 146:369–381.
