Abstract
The complete mitochondrial genome (mtgenome) sequences of Anopheles aconitus and A. splendidus were sequenced and analyzed in this study. They are 15,359 bp and 15,362 bp long, respectively and contains 13 protein-coding genes (PCGs), 22 tRNA genes (tRNAs), 2 rRNA genes (rRNAs), and 1 AT-rich control region (CR). The codon UUA (Leu) are prodominarily used in all 13 PCGs. ATN is mainly used as the initiation codon in theses PCGs except for COI and ND5 genes, which use TCG and GTG as the initiation codon, respectively and TAA as termination codon except for COI, COII, COIII, and ND4 that use the incomplete termination codon T. All of the tRNAs have the typical clover-leaf structure except for tRNASer(AGN), which lost the dihydrouridine (DHU) arm. The CRs have the highest A + T content of 92.97 and 93.18% in these two species, respectively. The phylogenetic relationships of 20 species in the subgenus Cellia were constructed using Maximum Likelihood based on concatenated nucleotide sequences of 13 PCGs with the selected best-fit GTR + I+G model. The 20 species are clearly divided into four monophyletic series: Pyretophorus, Neomyzomyia, Neocellia, and Myzomyia, and the Neomyzomyia is basal to the other three series. The Neocellia and Myzomyia are suggested to be sister groups and the Pyretophorus is proposed to sister with Neocellia + Myzomyia. This study provides a basis for further study on mtgenome and phylogenetics in the subgenus Cellia.
Anopheles aconitus Dönitz (Dönitz Citation1902) belongs to the minimus subgroup of the Funestus Group, the Myzomyia series in the subgenus Cellia. It is broadly distributed from Sri Lanka, southern and eastern India and southern Nepal, eastward to southern China (Hainan and Yunnan Province), south into Indonesia as far east as Babar Island in the southern Maluku archipelago (Harrison Citation1980; Sinka et al. Citation2011; Chen et al. Citation2012). It has been affirmed as a vector of malaria parasites in Thailand and is considered to be the primary malaria vector in Malaysia and Indonesia (Gould et al. Citation1965; Damar et al. Citation1981; Rahman et al. Citation1993; Barcus et al. Citation2002). Besides malarial parasites, Wuchereria bancrofti filariaes have also been found in this species in Flores, Indonesia (Atmosoedjono and Dennis Citation1977). Anopheles splendidus Koidzmi, belonging to the Jamesii Group of Neocellia Series, is widely distributed in southern and southeast of China, Southeast Asia, Afghanistan, Nepal, and Pakistan. It has been considered to be the vector of malaria and bancroftian filariasis in Hong Kong and Taiwan (Lu Citation1997). The mitochondrial fragments of An. aconitus and An. splendidus, such as the cytochrome-oxidase subunit 1 (COI), and the cytochrome-oxidase subunit 2 (COII) have been applied to the studies of molecular identification, phylogenetic and population genetics of these two species (Chen et al. Citation2012). However, the complete mtgenome sequences of the two species have not been reported. In this study, we report the complete mtgenomes of An. aconitus and An. splendidus, comparatively analyze the characteristics of the two species of mtgenomes and construct and discuss the phylogenetic relationships of all 20 known mtgenomes in the subgenus Cellia using nucleotide sequences of the 13 PCGs. This comprehensive study of the known mtgenomes in the subgenus Cellia establishes an information frame of subgenus Cellia mtgenomes and phylogenetics.
Adult individuals of An. aconitus and An. splendidus were collected from the Ja Htu Kong village (24°70.614N, 97°56.625E) in the Kachin Region of Myanmar (the border of Yunnan Province in China) in April 2012. Individual specimens were preserved in 80% ethanol after identification based on morphological characteristics (Dong Citation2010) and then stored at –80 °C before the DNA extraction. Total DNA was extracted from the individual adult mosquito and stored at –80 °C refrigerator in Institute of Entomology and Molecular Biology, Chongqing Normal University, using the DNeasy Blood and Tissue kit (250) (Qiagen 69506, Duesseldorf, Germany) (Yang et al. Citation2014). The complete mtgenome of the two species were amplified using 18 primer pairs designed by Zhang et al. (Citation2013) and the PCRs were carried out as described in Hua et al. (Citation2016). The resultant amplified products were verified by electrophoresis on a 1% agarose gel and were sequenced on ABI 3730XL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). All fragments were sequenced in both directions.
The complete mtgenomes of An. aconitus and An. splendidus are 15,359 bp (GenBank number KX887320) and 15,362 bp (GenBank number KX887321) in length, respectively. They are both comprised of 37 typical mtgenome genes (13 PCGs, 22 tRNAs, and 2 rRNAs) and a non-coding region. The 22 genes (9 PCGs and 13 tRNAs) are located on the heavy coding strand (H strand), while the other 15 genes (4 PCGs, 9 tRNAs, and 2 rRNAs) are on the light strand (L strand). Seven and nine intergenic spacers exist in the An. aconitus and An. splendidus mtgenome, which have a total length of 44 bp and 43 bp ranging from 1 bp to 17 bp each, respectively; and the longest intergenic spacer is located between tRNASer(UCN) and ND1. In addition, the 13 PCGs are overlapped with a total length of 29 bp and 30 bp in both An. aconitus and An. splendidus mtgenomes, with the overlaps ranging from 1 bp to 7 bp and the longest two spacers are located between ATP8 and ATP6, and ND4 and ND4L in these two species. The mtgenomes of An. aconitus showed a high nucleotide bias with 78.25% of AT and 21.75% of GC (39.94% A; 38.31% T; 9.19% G; and 12.56% C). The overall nucleotide composition of An. splendidus is 40.18% A, 37.73% T, 9.41% G, and 12.56% C, and CR has the highest AT content (93.18%), The AT contents of rRNAs, tRNAs and PCGs in An. aconitus are 82.02, 78.78, and 76.77% and those of An. splendidus have similar values of 82.02, 78.74, and 76.3%. In the An. aconitus and An. splendidus PCGs, except for COI and ND5 that use TCG and GTG as a start codon, the start codons of most PCGs follow the typical ATN rules: five genes (COII, ATP6, COIII, ND4, and CytB) use ATG as start codon, three genes (ATP8, ND6, and ND1) use ATT, two genes (ND3 and ND4L) use ATA, and one gene (ND2) use ATC. The use of the GTG start codon has been documented for mtDNA-encoded genes in various organisms, including subgenus Cellia (Krzywinski et al. Citation2011). The termination codons are the complete termination codon TAA or incomplete termination codon T. Most PCGs have the complete termination codon TAA, whereas four genes (COI, COII, COIII, ND4) utilize the incomplete stop codon T, which could be completed as TAA via postranscriptional polyadenylation (Ojala et al. Citation1981). The 22 tRNAs of An. aconitus and An. splendidus are 1477 bp and 1489 bp long, respectively, with a range from 64 bp to 72 bp. The 20 tRNA genes of An. aconitus and 21 tRNA genes of An. splendidus were identified using the program tRNAscan-SE except for tRNASer(AGN) and tRNAArg in comparison with known mosquito mitochondrial sequences. All tRNAs can be folded into typical clover-leaf structure except for tRNASer(AGN), which lost the dihydrouridine (DHU) arm. The 12S rRNAs in the An. aconitus and An. splendidus mtgenomes are both located between trnV and A + T control region and the 16S rRNAs between trnL and trnV, as in the mtgenomes in other metazoan species (Beard et al. Citation1993). The 12S rRNAs are totally 794 bp long with AT contents 79.7 and 80.4%, respectively and the 16S rRNAs are 1325 bp and 1330 bp long with AT contents 83.4 and 83%, respectively in the two species. The control region of the An. aconitus and An. splendidus mtgenome are located between 12S rRNA and tRNAIle. The lengths are 526 bp and 514 bp, with the highest A + T content 92.97 and 93.18%, respectively.
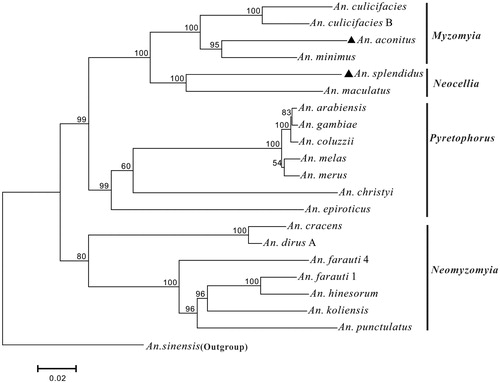
We constructed the phylogenetic relationship of mtgenomes of An. aconitus and An. splendidus and 18 other subgenus Cellia species using Maximum Likelihood (ML) method with Mega 7.0 and with the Anopheles sinensis mtgenome (MF322628) (Ding et al. Citation2018) as an outgroup. The nucleotide sequences of the 13 PCGs were used in the phylogenetic analysis and the best model GTR + I+G was selected using ModelTest for the analysis. The bootstrap values were calculated based on 1000 replicates and the bootstrap values larger than 50% are noted on the corresponding nodes of the phylogenetic tree (). The 18 other subgenus Cellia species of mtgenome sequences were downloaded from GenBank. The result shows that the newly sequenced An. aconitus and An. splendidus are grouped into Myzomyia Series and Neocellia Series, respectively as expected. The 20 species in the subgenus Cellia are clearly divided into four monophyletic Series: Pyretophorus, Neomyzomyia, Neocellia, and Myzomyia with at least 99% bootstrap values of support. The Neomyzomyia Series is basal to other three Series. The Neocellia Series and Myzomyia Series are suggested to be sister groups and the Pyretophorus Series is proposed to sister with Neocellia + Myzomyia clade. These results are consistent with the earlier phylogenetic study result based on morphological and molecular data (Harbach and Kitching Citation2005; Harbach Citation2013).
Figure 1. The phylogenetic relationships of 20 species in the subgenus Cellia constructed by maximum likelihood based on nucleotide sequences of 13 PCGs. The bootstrap values larger than 50% are denoted on the corresponding nodes. Species names with GenBank mitogenome accession numbers in bracket: An. culicifacies (NC028216), An. culicifacies B (NC027502), An. aconitus (KX887320), An. minimus (KT895423), An. splendidus (KX887321), An. maculatus (NC028218), An. arabiensis (NC028212), An. gambiae (NC002084), An. coluzzii (NC028215), An. melas (NC028219), An. merus (NC028220), An. christyi (NC028214), An. epiroticus (NC028217), An. cracens (JX219733), An. dirus A (JX219731), An. farauti 4 (JX219735), An. farauti 1 (JX219741), An. hinesorum (JX219734), An. koliensis (JX219743), An. punctulatus (JX219738), An. sinensis (MF322628).
Author contributions
Conceived and designed the research: BC, ZTY. Performed the experiments: ZTY, WBF. Analyzed the data and wrote the paper: ZTY, BC.
Disclosure statement
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the paper. It is stated that research meets ethical guidelines.
Additional information
Funding
References
- Atmosoedjono S, Dennis DT. 1977. Anopheles aconitus and Anopheles subpictus naturally infected with wuchereria bancrofti in Flores, Indonesia. Mos News. 37:529.
- Barcus MJ, Laihad F, Sururi M, Sismadi P, Marwoto H, Bangs MJ, Baird JK. 2002. Epidemic malaria in the Menoreh Hills of Central Java. Am J Trop Med Hyg. 66:287–292.
- Beard CB, Hamm D, Collins FH. 1993. The mitogenome of the mosquito Anopheles gambiae: DNA sequence, genome organization, and comparisons with mitochondrial sequences of other insects. Insect Mol Biol. 2:103–124.
- Chen B, Harbach RE, Walton C, He ZB, Zhong DB, Yan GY, Butlin RK. 2012. Population genetics of the malaria vector Anopheles aconitus in China and Southeast Asia. Infect Genet Evol. 12:1958–1967.
- Damar T, Fleming GA, Gandahusada S, Bang YH. 1981. Nocturnal indoor resting heights of the malaria vector Anopheles aconitus and other Anopheles (Diptera: Culicidae) in Central Java, Indonesia. J Med Ent. 18:362–365.
- Ding YR, Li B, Zhang YJ, Mao QM, Chen B. 2018. Complete mitogenome of Anopheles sinensis and mitochondrial insertion segments in the nuclear genomes of 19 mosquito species. PLoS ONE. 13:e0204667.
- Dong XS. 2010. The mosquito fauna of Yunnan China, volume one [M]. Yunnan: Yunnan Science & Technology Press; p. 1–394.
- Dönitz W. 1902. Beitrage zur Kenntniss der Anopheles. Z Hyg Infektionskr. 41:15–88.
- Gould DJ, Esah S, Pranith U. 1965. Relation of Anopheles aconitus to malaria transmission in the central plain of Thailand. Trans Roy Soc Trop Med Hyg. 59:441–442.
- Harbach RE. 2013. Anopheles mosquitoes - New insights into malaria vectors. Chapter 1: The Phylogeny and classification of Anopheles [M]. UK: InTechOpen; p. 1–55.
- Harbach RE, Kitching IJ. 2005. Reconsideration of Anopheline phylogeny (Diptera: Culicidae: Anophelinae) based on morphological data. Syst Biodivers. 12:345–374.
- Harrison BA. 1980. Medical entomology studies-XI II. The Myzomyia series of Anopheles (Cellia) in Thailand, with emphasis on intra-interspecific variations (Diptera: Culicidae). Contrib Am Entomol Instit. 17:1–195.
- Hua YQ, Ding YR, Yan ZT, Si FL, Luo QC, Chen B. 2016. The complete mitochondrial genome of Anopheles minimus (Diptera: Culicidae) and the phylogenetics of known Anopheles mitogenomes. Insect Sci. 23:353–365.1
- Krzywinski J, Li C, Morris M, Conn JE, Lima JB, Povoa MM, Wilkerson RC. 2011. Analysis of the evolutionary forces shaping mitogenomes of a Neotropical malaria vector complex. Mol Phylogenet Evol. 58:469–477.
- Lu BL. 1997. Fauna Sinica. Insecta. Diptera: Culicidae 1. Vol. 8. China: Science Press.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Rahman WA, Hassan AA, Adanan CR. 1993. Seasonality of Anopheles aconitus mosquitoes, a secondary vector of malaria, in an endemic village near the Malaysia-Thailand border. Acta Tropica. 55:263–265.
- Sinka ME, Bangs MJ, Manguin S, Chareonviriyaphap T, Patil AP, Temperley WH, Gething PW, Elyazar IR, Kabaria CR, Harbach RF, et al. 2011. The dominant Anopheles vector of human malaria in the Asia-Pacific region: occurrence data, distribution maps and bionomic precis. Parsit Vectors. 4:89.
- Yang FL, Li XD, Yan ZT, Chen B. 2014. The molecular identification markers of Anopheles sinensis. J Chongqing Normal Univ (Nat Sci). 31:40–44.
- Zhang NX, Zhang YJ, Yu G, Chen B. 2013. Structure characteristics of the mitochondrial genomes of Diptera and design and application of universal primers for their sequencing. Acta Entomol Sinica. 56:398–407.
