Abstract
Ammopiptanthus mongolicus is a rare and Endangered plant of Ammopiptanthus (Papilionoideae) in China. It grows mainly in the arid and semi-arid regions. The complete chloroplast (cp) genome was assembled by Illumina paired-end reads data. The circular cp genome is 153,997 bp in size, including a large single copy (LSC) region of 83,957 bp, a small single copy (SSC) region of 18,008 bp and a pair of inverted repeat (IRs) regions of 26,016 bp. Besides, 5 genes possess a single intron, while another three genes ycf3, rps12, and clp have a couple of introns. The GC content of the entire A. mongolicus cp genome, LSC, SSC, and IR regions are 36.9, 34.6, 30.5, and 42.8%, respectively. Based on the concatenated coding sequences of cp PCGs, the phylogenetic analysis showed that A. mongolicus and A. nanus are closely related to each other within the family Leguminosae.
Ammopiptanthus mongolicus belongs to the family Papilionoideae, which is a traditional medicinal plant in China (Fu Citation1991). It grows mainly in the arid and semi-arid regions of Ningxia, Xinjiang, Inner Mongolia and so on (Liu et al., Citation1995). Previous studies had shown that the stem and leaf of the plant have commonly been used in traditional medicine, which was recorded as a famous and historic drug for its effects of flipping wind dehumidification and relieving muscle stasis. As an only evergreen broad-leaf shrub in the northwest desert of China, A. mongolicus shows very strong resistance to both drought and cold stresses (Huang et al. Citation2012).
The conserved quadripartite structure of a typical circular cp genome include a pair of inverted repeats (IRs), separated by a large single-copy region (LSC) and a small single-copy region (SSC) (Wolfe et al. Citation1992; Lee et al. Citation2007). This study will be very useful for the research on the phylogenetic relationships of A. mongolicus and Papilionoideae.
Fresh leaves of A. mongolicus were collected in the Yinchuan Botanical Garden (38°28′N, 106°16′E; Ningxia, NW China). A voucher specimen (P02C) is deposited at the State Key Laboratory Seedling Bio-engineering in Ningxia Forestry Institute. We used the modified CTAB method to extract the total genomic DNA (Doyle and Doyle Citation1987). A shotgun library constructed following the manufacturer’s protocol for the Illumina HiSeq X Ten Sequencing System (Illumina, San Diego, CA, USA). We assembled the cp genome using the program MITObim v1.8 (DSM Nutritional Products Ltd, Kaiseraugst, Switzerland) (Hahn et al. Citation2013), with that of Ammopiptanthus nanus (GenBank: KY034454) as the initial reference. The web-based tool OGDRaw v1.2 (http://ogdraw.mpimp-golm.mpg.de/) was employed to generate a map of the complete cp genome (Lohse et al. Citation2013). The complete cp genome sequence has been submitted to GenBank (accession number MK704436).
The complete cp genome is a circular and double-stranded DNA molecule with a typical quadripartite structure, containing two inverted repeat (IRs) regions, a large single copy (LSC) region, and a small single copy (SSC) region. In all, we received 23,776,174 raw paired-end reads, which length distribution in 153,997 bp (GC, 36.9%), the length of LSC region, IRs regions and SSC region in 83,957 (GC, 34.6%), 26,016 (42.8%), and 18,008 (30.5%), respectively.
It encodes 130 complete genes, including 84 protein-coding genes, 37 transfer RNA genes, and 8 ribosomal RNA genes. In addition, 5 PCG genes (atpF, ndhA, ndhB, rpl2, and rpoC1) possess a single intron, 76 PCG genes no intron, 3 other genes (clpP, rps12, and ycf3) harbor two introns, 5 tRNA genes (trnA-UGC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) harbor a single intron.
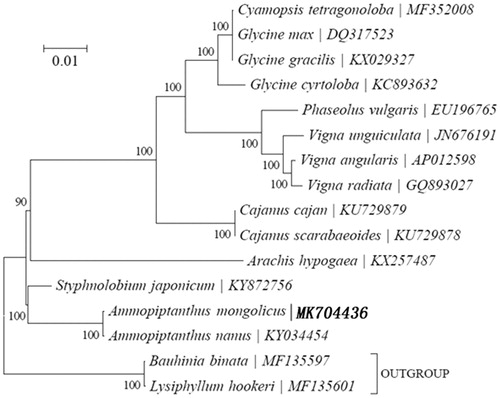
To investigate the phylogenetic position of A. mongolicus, we constructed a neighbour-joining (NJ) phylogenetic tree () based on the concatenated coding sequences of 17 chloroplastp PCGs for 17 plastid genomes from published species of Compositae using MEGA7 with 1000 bootstrap replicates (Kumar et al. Citation2016) (http://www.megasoftware.net/). From the phylogenetic tree analysis, we find that A. mongolicus and Ammopiptanthus nanus (KY034454) are closely related to each other within the family Papilionoideae.
Disclosure statement
The authors report no conflicts of interest and are solely responsible for the content and writing of this paper.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fu LG. 1991. China plant red data book: rare and Endangered plants. Vol. 1. Beijing: Sciences Press.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Huang G-T, Ma S-L, Bai L-P, Zhang L, Ma H, Jia P, Liu J, Zhong M, Guo Z-F. 2012. Signal transduction during cold, salt, and drought stresses in plants. Mol Biol Rep. 39:969–987.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lee HL, Jansen RK, Chumley TW, Kim KJ. 2007. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol Biol Evol. 24:1161–1180.
- Liu, JQ, Qiu, MX, Shi, QH. 1995. Studies on the community of Holly in sand. China Desert. 15:109–115.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucl Acids Res. 41:W575–W81.
- Wolfe KH, Morden CW, Ems SC, Palmer JD. 1992. Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant: loss or accelerated sequence evolution of tRNA and ribosomal protein genes. J Mol Evol. 35:304–317.