Abstract
In this paper, we characterized the complete mitogenomes of Aratinga solstitialis, Pionites leucogaster, and Poicephalus senegalus through Sanger sequencing method. The full lengths of the three mitogenomes are 16,986 bp, 18,858 bp, and 18,022 bp, respectively. All genes exhibit original gene arrangement in A. solstitialis mitogenome, but gene rearrangements occur in P. leucogaster and P. senegalus mitogenomes. Bayesian tree indicated that the three parrots were not closely related to each other. The results obtained here can contribute to phylogenetic analysis and biological conservation of parrots further.
According to statistics, there are about 400 species in the Psittaciformes (Parrot) around the world, but many of them are threatened with extinction (Olah et al. Citation2016; Heinsohn et al. Citation2018). Aratinga solstitialis is distributed in the semideciduous forests and savannas of north-eastern Roraima, Brazil, and adjacent Guyana (BirdLife International Citation2016a). Pionites leucogaster is only found in the lowland tropical rainforests from the Rio Madeira to Maranhão, Brazil (BirdLife International Citation2016b). Poicephalus senegalus is widely distributed in the forest and savanna of West African countries (BirdLife International Citation2016c). The wild populations of A. solstitialis and P. leucogaster are still decreasing, mainly due to the habitat loss and biological resource use. They have been listed as Endangered on the IUCN Red List of Threatened Species (BirdLife International Citation2016a, Citation2016b). However, the wild population of P. senegalus is so large and stable that it is listed as a species of least concern by IUCN (BirdLife International Citation2016c). Here, we characterized and compared the complete mitogenomes of the three parrots.
Captive-bred specimens of the three parrots were collected from Nanjing Hongshan Forest Zoo (N32°09′, E118°80′) and stored in College of Biology and the Environment, Nanjing Forestry University (Accessions HSZ201901-HSZ201903). DNA extraction, PCR reaction and product purification were carried out by the DNAiso reagent, Ex Taq and Minibest agarose gel DNA extraction kit, respectively (Takara, Beijing, China). Genome information was obtained through Sanger sequencing.
The complete mitogenomes of A. solstitialis (GenBank accession MK343132), P. leucogaster (GenBank accession MK759905) and P. senegalus (GenBank accession MK749396) were 16,986 bp, 18,858 bp, and 18,022 bp in length, respectively. All mitogenomes were AT-rich, with the following nucleotide compositions: 23.8% T, 31.9% C, 30.3% A, 14.0% G for A. solstitialis; 23.5% T, 32.4% C, 30.0% A, 14.0% G for P. leucogaster; 22.8% T, 33.0% C, 29.3% A, 14.9% G for P. senegalus. The data was in accordance with the nucleotide composition in the avian mitogenome (Eberhard and Wright Citation2016; Liu et al. Citation2019). The mitogenome of A. solstitialis included 22 tRNA genes, 2 rRNA genes, 13 protein-coding genes, and a control region. Besides above components, there was another control region in P. leucogaster and P. senegalus, respectively.
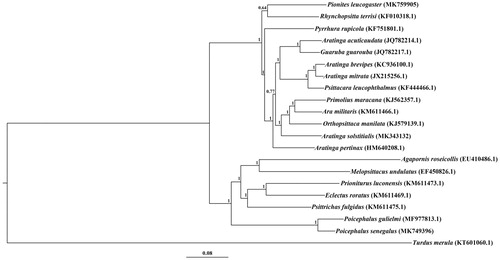
To assess the phylogenetic positions of A. solstitialis, P. leucogaster, and P. senegalus, Bayesian tree was reconstructed based on complete mitogenomes of 20 parrot species using Turdus merula as an outgroup (Huelsenbeck et al. Citation2001). The phylogenetic tree indicated that the three parrots were not closely related to each other (). P. leucogaster was closely related to Rhynchopsitta terrisi (). P. senegalus was placed as sister to Poicephalus gulielmi of the same genus (). However, unexpectedly, A. solstitialis did not cluster with other species of the same genus, suggesting the phylogenetic relationship among the species in Aratinga genus was not well solved (). The complete mitogenomes obtained here provide important DNA data for phylogenetic analysis and biological conservation of parrots in further.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- BirdLife International. 2016a. Aratinga solstitialis. The IUCN Red List of Threatened Species 2016:e.T62233372A95192947. http://dx.doi.org/10.2305/IUCN.UK.2016-3.RLTS.T62233372A95192947.en. Downloaded on 03 May 2019.
- BirdLife International. 2016b. Pionites leucogaster. The IUCN Red List of Threatened Species 2016:e.T62181308A95191855. http://dx.doi.org/10.2305/IUCN.UK.2016-3.RLTS.T62181308A95191855.en. Downloaded on 03 May 2019.
- BirdLife International. 2016c. Poicephalus senegalus. The IUCN Red List of Threatened Species 2016:e.T22685295A93066490. http://dx.doi.org/10.2305/IUCN.UK.2016-3.RLTS.T22685295A93066490.en. Downloaded on 03 May 2019.
- Eberhard JR, Wright TF. 2016. Rearrangement and evolution of mitochondrial genomes in parrots. Mol Phylogenet Evol. 94:34–46.
- Heinsohn R, Buchanan KL, Joseph L. 2018. Parrots move to centre stage in conservation and evolution. EMU. 118:1–6.
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. 2001. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 294:2310–2314.
- Liu HY, Ding YF, Chen R, Tong Q, Chen PX. 2019. Complete mitogenomes of two Psittacula species, P. derbiana and P. eupatria. Mitochondrial DNA B. 4:692–693.
- Olah G, Butchart SHM, Symes A, Medina Guzmán I, Cunningham R, Brightsmith DJ, Heinsohn R. 2016. Ecological and socio-economic factors affecting extinction risk in parrots. Biodivers Conserv. 25:205–223.