Abstract
Yushu yak (Bos grunniens) is a local breed of yaks with strong adaptation to the high-altitude, cold, and anoxic environments. In this study, its complete mitochondrial genome was assembled from Illumina sequencing data. The genome was determined to be 16,324 bp in size with a moderately asymmetric nucleotide composition (61.0% A + T) and encodes the typical set of 37 mitochondrial genes. The PCGs are initiated with the typical ATA or ATG codons and are terminated with TAA, TAG, or the incomplete stop codon T––. Phylogenetic analysis suggests that Yushu yak is most closely related to Qinghai Plateau yak.
Domestic yaks (Bos grunniens) are an iconic symbol of the Qinghai-Tibetan Plateau and adjacent regions and provide meat, milk, transportation, and other necessities for local residents (Qiu et al. Citation2012). To date, multiple local breeds have been cultivated across the whole distribution range (Chu et al. Citation2016; Guo et al. Citation2016; Wu, Chu, et al. Citation2016; Wu, Ding, et al. Citation2016). Here, we report the complete mitochondrial genome (GenBank accession: MK704512) for Yushu yak, a local breed of yaks with its distribution mostly restricted to Qumalai County, Yushu Tibetan Autonomous Prefecture, Qinghai Province, China. This breed is characterized by its strong adaptation to the high-altitude, cold and anoxic environments.
A blood sample of Yushu yak was collected from Qumalai County, Yushu Tibetan Autonomous Prefecture, Qinghai Province (34°13'66''N, 95°81'72''E). A voucher specimen is deposited in the Key Laboratory of Yak Breeding Engineering of Gansu Province, Lanzhou Institute of Husbandry and Pharmaceutical Sciences (Lanzhou, Gansu Province, China). Total genomic DNAs were extracted using the QIAamp DNA Blood Mini Kit (Qiagen, CA, USA). Following the library preparation, high-throughput sequencing was conducted with the Illumina HiSeq X™ Ten Sequencing System (Illumina, CA, USA) by Annoroad Gene Technology (Beijing, China). In all, 15.1 M raw reads of 150 bp were generated, and were used for mitogenome assembly with MITObim v1.9 (Hahn et al. Citation2013); the reference sequence (JQ692071) was retrieved from a previously published study (Qiu et al. Citation2012). The yielded mitogenome sequence was annotated by aligning with those of its congeners, followed by a double-check with the prediction of the MITOS web server (Bernt et al. Citation2013).
The mitochondrial genome of Yushu yak (MK704512) is 16,324 bp long with a moderately asymmetric base composition (33.7% A, 25.8% C, 13.2% G & 27.3% T; ‘light strand’). It harbours the typical set of 37 animal mitochondrial genes (13 protein-coding genes, 22 tRNAs, and two rRNAs) and one non-coding control region. The PCGs are initiated with the typical ATA (ND2, ND3, and ND5) or ATG (the 10 others) codons. Three types of stop codons are annotated, including TAG (ND2), the incomplete stop codon T–– (COX3, ND3, and ND4), and TAA (the nine others). The tRNAs range in size from 60 (tRNA-SerAGN) to 75 bp (tRNA-LeuUUR) with a total length of 1509 bp. The two rRNAs are 957 bp (12S rRNA) and 1571 bp (16S rRNA) in length, respectively, and are separated by tRNA-Val. The 894-bp-long control region is located between tRNA-Pro and tRNA-Phe. There are 15 intergenic spacer and nine intergenic overlapping regions across the genome with a total length of 61 and 75 bp, respectively.
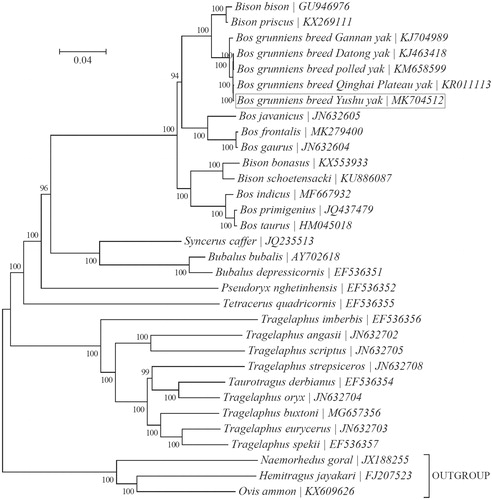
To investigate its relationship with other yak breeds and consubfamilial species, a Bayesian phylogeny was reconstructed using the concatenated sequences of all 13 protein-coding genes with MrBayes v3.1.1 (Ronquist and Huelsenbeck Citation2003) as implemented in TOPALi v2.5 (Milne et al. Citation2009) (). Phylogenetic analysis suggests that Yushu yak is more closely related to Qinghai Plateau yak than to three other local breeds (Gannan, Datong, and polled yaks).
Figure 1. Phylogeny of the subfamily Bovinae based on the Bayesian analysis of the concatenated sequences of 13 mitochondrial protein-coding genes (alignment size: 10,593 bp). The best-fit nucleotide substitution model is ‘GTR + G+I’. The support values are placed next to the nodes. Three species within the subfamily Caprinae were included as outgroup taxa.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Chu M, Wu X, Liang C, Pei J, Ding X, Guo X, Bao P, Yan P. 2016. The complete sequence of mitochondrial genome of polled yak (Bos grunniens). Mitochondr DNA A. 27:2032–2033.
- Guo X, Pei J, Bao P, Chu M, Wu X, Ding X, Yan P. 2016. The complete mitochondrial genome of the Qinghai Plateau yak Bos grunniens (Cetartiodactyla: Bovidae). Mitochondr DNA A. 27:2889–2890.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 25:126–127.
- Qiu Q, Zhang G, Ma T, Qian W, Wang J, Ye Z, Cao C, Hu Q, Kim J, Larkin DM, et al. 2012. The yak genome and adaptation to life at high altitude. Nat Genet. 44:946–949.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Wu X, Chu M, Liang C, Ding X, Guo X, Bao P, Yan P. 2016. The complete mitochondrial genome sequence of the Datong yak (Bos grunniens). Mitochondr DNA A. 27:433–434.
- Wu X, Ding X, Chu M, Guo X, Bao P, Liang C, Yan P. 2016. Characterization of the complete mitochondrial genome sequence of Gannan yak (Bos grunniens). Mitochondr DNA A. 27:1014–1015.
