Abstract
The complete mitogenome of Myersina filifer was sequenced, which is a closed double-stranded circular molecule of 16,505 bp, we analyzed the main features in terms of the genome organization, gene arrangement, and codon usage. The overall base composition includes A(27.4%), C(28.0%), T(27.1%) and G(17.5%). Moreover, the 13 protein-coding genes (PCGs) encode 3801 amino acids in total, 12 of which use the initiation codon ATG except COI uses GTG, most of them have a complete stop codon, whereas four PCGs end with the incomplete stop codon TA– or a single T. The phylogenetic tree based on the Neighbor-Joining method was conducted to provide relationship within Gobiidae, the NJ tree demonstrated that M. filifer has the closest relationship with M. macrostoma, which contributed to phylogenetic studies of Gobiidae and additional information of management for this species.
Myersina is a genus of ray-finned fish from the family Gobiidae, the true gobies which are found from the Atlantic coast of South Africa through the Indian Ocean to the western Pacific Ocean (Winterbottom Citation2002). There are currently nine recognized species in this genus, while only one Myersina mitogenome (M. macrostoma) has been completely sequenced, denser species sampling has been demonstrated to facilitate phylogenetic analysis (Wang et al. Citation2001; Jin et al. Citation2013). Herein, we sequenced the complete mitochondrial genome of M. filifer, its molecular features are described in detail. This study was expected to improve the utility of Myersina mitogenomes and lay the foundation for further phylogenetic studies on M. filifer and related species.
Specimens of M. filifer were sampled by commercial bottom trawling in the South China Sea (16°48′42″N; 112°22′25″E) and stored in the laboratory of Zhejiang Ocean University with accession number 201804YS. The total genomic DNA was extracted using the phenol-chloroform method (Köchl et al. Citation2005). The complete mitochondrial genome of M. filifer is 16,511 bp in length (GenBank Accession Number: MK758112), consisting of 13 PCGs, 22 tRNAs, 2 rRNAs, 1 replication origin, and a control region, this feature is similar to the typical mitogenome of other vertebrates (Cheng et al. Citation2012; Prosdocimi et al. Citation2012). The overall base composition is 27.4, 28.0, 17.4, and 27.1% for A, C, G, and T, respectively, with a slight AT bias 55.5%. Excluding termination codons, 13 PCGs encode 3801 amino acids in total. All PCGs use the initiation codon ATG except for COI uses GTG, most of them have TAA or TAG as the stop codon, whereas COIII gene uses TA and three PCGs (COII, ND4, and Cytb) use a single T as the termination codons, these incomplete termination codons are presumably completed as TAA via post-transcriptional polyadenylation (Ojala et al. Citation1981). The 12S and 16S genes are 947 bp and 1682 bp, respectively. Twenty-two tRNA genes are interspersed between rRNAs and PCGs and their secondary structures were generated using the program tRNAs-can-SE (Lowe and Eddy Citation1997), all of them can fold into a typical cloverleaf structure with an exception of tRNA-Ser (AGN), which lacks a dihydrouridine arm. The OL is located in a cluster of five tRNA genes (WANCY), which has the potential to fold into a stable stem-loop secondary structure, with a stem formed by 12 paired nucleotides and a loop of 12 nucleotides; The control region is 858 bp long, the core sequence of the terminal-associated sequence (ACATATATG) was identified in the CR of M. filifer, which is identical to that in other teleostean mitogenomes (Zhang et al. Citation2003).
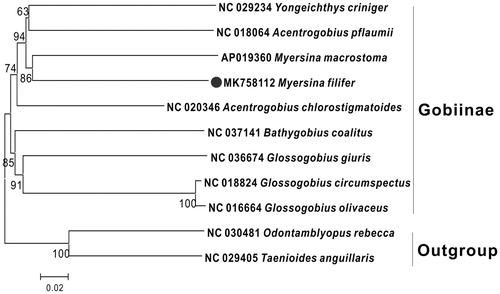
To explore the phylogenetic position of this M. filifer, we used MEGA7 (Kumar et al. Citation2016) to construct a phylogenetic tree based on the NJ analysis. The result of the present study supports M. filifer has the closest relationship with M. macrostoma (). It is critical to explore more mt genomic data to better understand the mt genome of M. filifer and reconstruct the phylogenetic relationship within Gobiidae.
Figure 1. Neighbor Joining (NJ) tree of 9 Gobiidae species based on 12 PCGs. The bootstrap values are based on 1000 resamplings. The number at each node is the bootstrap probability. The number before the species name is the GenBank accession number. The genome sequence in this study is labeled with a black spot.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Cheng Y, Wang R, Sun Y, Xu T. 2012. The complete mitochondrial genome of the small yellow croaker and partitioned Bayesian analysis of Sciaenidae fish phylogeny. Genet Mol Biol. 35:191–199.
- Jin X, Zhao S, Sun Y. 2013. Complete mitochondrial genome sequence and structure of control region of the Amoya chusanensis (Perciformes, Gobioidei). Mitochondr DNA. 24:403–405.
- Köchl S, Niederstätter H, Parson W. 2005. DNA extraction and quantitation of forensic samples using the phenol-chloroform method and real-time PCR. Methods Mol Biol. 297:13.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Ojala D, Montoya J, Attardi G. 1981. TRNA punctuation model of RNA processing in human mitochondrial. Nature. 290:470–474.
- Prosdocimi F, de Carvalho DC, de Almeida RN, Beheregaray LB. 2012. The complete mitochondrial genome of two recently derived species of the fish genus Nannoperca (Perciformes, Percichthyidae). Mol Biol Rep. 39:2767–2772.
- Wang HY, Tsai MP, Dean J, Lee SC. 2001. Molecular phylogeny of gobioid fishes (Perciformes: Gobioidei) based on mitochondrial 12S rRNA sequences. Mol Phylogenet Evol. 20:390–408.
- Winterbottom R. 2002. A redescription of Cryptocentrus crocatus Wongratana, a redefinition of Myersina Herre (Acanthopterygii; Gobiidae), a key to the species, and comments on relationships. Ichthyol Res. 49:69–75.
- Zhang P, Chen YQ, Zhou H, Wang XL, Qu LH. 2003. The complete mitochondrial genome of a relic salamander, Ranodon sibiricus (Amphibia: Caudata) and implications for amphibian phylogeny. Mol Phylogenet Evol. 28:620–626.
