Abstract
We reported and characterized the complete nucleotide sequence of the Spathiphyllum cannifolium chloroplast (cp) genome in this study. The circular molecule of the cp genome is 171,420 bp containing a pair of 31,460 bp IR regions, separated by a small single-copy region (SSC) of 16,331 bp and a large single-copy region (LSC) of 92,169 bp, respectively. A total of 132 genes were successfully annotated in the cp genome, including 87 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The G + C content of the whole cp genome is 35.62%, while the LSC, SSC, and IR region are 33.94, 28.02, and 40.05%, respectively.
Spathiphyllum cannifolium is an important tropical ornamental potted plant. Spathiphyllum cannifolium used in this paper was stored at the resource garden of environmental horticulture research institute, Guangdong Academy of agricultural sciences, Guangzhou, China.
We first extracted the cp genome DNA from young leaves of S. cannifolium (Shi et al. Citation2012) and second, shotgun libraries were constructed according to TruSeq™ DNA Sample Prep Kit for Illumina (San Diego, CA, USA). Third, the whole genome sequences were executed using Illumina Hiseq 4000 platform. Then, the sequences obtained were assembled using SOAP denovo (version 2.04) (Luo et al. Citation2012). The 39,877,869 filtered reads were assembled into the cp genome by SOAP de novo software and subsequent analysis were done by Celera Assembler (version 8.0, Rockville, MD, USA). The complete cp genome sequence has been submitted to Genbank with the accession number of MK372232.
The complete cp genome sequence of S. cannifolium is 171,420 bp in length, containing a pair of 31,460 bp IR regains, separated by large and small single-copy regions of 92,169 bp and 16,331 bp, respectively. The location of the LSC, IRa, SSC, and IRb is 1–92,169 bp, 92,170–123,629 bp, 123,630–139,960 bp and 139,961–171,420 bp, respectively. Overall, A content of the genome was 31.71%, while the other T, C, and G contents were 32.67, 18.10, and 17.52%, respectively. LSC contains 62 protein-coding genes and 22 tRNAs, while the SSC contains only 1 tRNA and 11 protein-coding genes. IR region includes 7 protein-coding genes, 7tRNAs, and 4 rRNAs. Among these identified genes, a single intron located in 15 different genes (trnK-UUU, trnG-GCC, trnL-UAA, trnV-UAC, ttrnI-GAU, trnA-UGC, rps16, rpoC1, petB, petD, rpl16, rpl2, ndhB, ycf68, and ndhA), and two introns located in ycf3 and clpP. The rps12 gene is a trans-spliced gene with the 5′ end located in the LSC region and the duplicated 3′ ends in the IR regions.
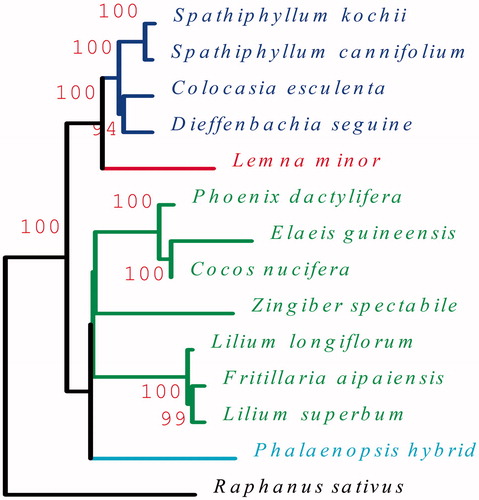
Phylogenetic relationships were constructed by a set of 24 protein-coding genes including atpB, atpE, atpF, atpH, atpI, ccsA, cemA, matK, petA, petG, petN, psaA, psaB, psaC, psbC, psbD, psbE, psbF, psbI, psbJ, psbK, psbN, psbZ, and rps15 from 14 chloroplast genomes. Sequence alignment and maximum likelihood (ML) analysis were performed by DNAMAN (Version 8.0). Spathiphyllum cannifolium was situated as the sister of most closely related to S. kochii in the Araceae (). This study will be useful for further analysis of genetic diversity, molecular markers, and molecular breeding.
Figure 1. The ML phylogenetic tree of S. cannifolium and other species based on 24 protein-coding genes from 14 plant taxa. Raphanus sativus (NC_024469) was set as outgroup. Genbank accession numbers for each plant are: Colocasia esculenta (NC_016753), Dieffenbachia seguine (NC_027272), Spathiphyllum cannifolium (MK372232), Spathiphyllum kochii (KR270822), Lemna minor (NC_010109), Phalaenopsis hybrid (NC_007499), Cocos nucifera (KF285453), Elaeis guineensis (JF274081), Phoenix dactylifera (GU811709), Lilium longiflorum (KC968977), Lilium superbum (NC_026787), Fritillaria taipaiensis (NC_023247), Zingiber spectabile (NC_020363).
Author contributions
Performed the experiments investigation, project administration, writing the original draft and data curation: XL.
Prepared the resources: DL.
Supervised the project and made revisions to the manuscript: GZ XW.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Shi C, Hu N, Huang H, Gao J, Zhao YJ, Gao LZ. 2012. An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. PLoS One. 7:e31468.
