Abstract
Riccia fluitans L. (Marchantiales) is an aquatic liverwort usually found in lentic environments. Here, we presented a complete chloroplast genome of R. fluitans (MK645896) which is 121,682 bp long and has four subregions: 81,626 bp of large single copy (LSC) and 20,254 bp of small single copy (SSC) regions are separated by 9,901 bp of inverted repeat (IR) regions including 131 genes (87 protein-coding genes, 8 rRNAs, and 36 tRNAs). The overall GC content is 29.0% and those in the LSC, SSC, and IR regions are 26.6%, 24.6%, and 42.9%, respectively. Phylogenetic trees show incongruence with Bryophyte taxonomy, requiring intensive researches to find genes causing this incongruence.
Genus Riccia L. covers approximately 150 species, which is the most diverse genus in order Marchantiales (Ayub et al. Citation2014). Riccia fluitans L, one of the most common species in Riccia, is distributed world widely (Manju et al. Citation2012). It is a conspicuous aquatic liverwort usually found in lentic environments, characterized by floating thalli which regularly fork into equal branches; however, it is difficult to be distinguished with other species in R. fluitans complex (Berrie Citation1964). It can also grow in both aquatic and terrestrial habitats with various morphological types along with different habitats (Carter Citation1935). Recent phylogenetic study presented that Ricciella (Braun) Boulay, the subgenus of R. fluitans, was polyphyletic (Cargill et al. Citation2016), requiring more information of molecular markers to clarify its phylogenetic position. We completed chloroplast genome of R. fluitans as a fourth representative species of Marchantiales families considering both chloroplast and mitochondrial genomes (Kwon, Kim, et al. Citation2019b, Citation2019a, Citation2019c; Kwon, Min, et al. Citation2019; Park et al. Citationaccepted.
Thallus of R. fluitans was collected in a marsh in Namyangju city, South Korea (Voucher in InfoBoss Cyber Herbarium (IN); W. Kwon, IB-50004) and its DNA was extracted by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeqX at Macrogen Inc., Korea, and de novo assembly and sequence conformation processes were done by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation based on Marchantia polymorpha subsp. ruderalis chloroplast genome (NC_037507).
Riccia fluitans chloroplast genome (Genbank accession is MK645896) is 121,682 bp (GC ratio is 29.0%) and has four subregions: 81,626 bp of large single copy (26.6%) and 20,254 bp of small single copy (24.6%) regions are separated by 9901 bp of the inverted repeat (IR; 42.9%). It contained 131 genes (87 protein-coding genes, 8 rRNAs, and 36 tRNAs); 9 genes (4 rRNAs and 5 tRNAs) are duplicated in IR regions.
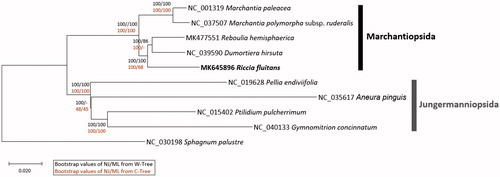
Ten complete chloroplast genomes including R. fluitans were used for constructing neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenic trees using MEGA X (Kumar et al. Citation2018) after aligning whole chloroplast genomes (named as W-Tree) and 63 conserved protein-coding genes (named as C-Tree) using MAFFT 7.388 (Katoh and Standley Citation2013). Phylogenetic trees show incongruent with previous phylogenetic study of Marchantiales (Crandall‐Stotler et al. Citation2016) by showing that Dumortiera hirsutais clustered with Reboulia hemisphaerica from W-Tree or D. hirsutais in the same clade of R. fluitans from C-Tree (). This phenomenon has also been observed in ant species (Park et al. Citation2019b, Citation2019a). In addition, maximum likelihood tree is also incongruent with neighbor joining tree in this clade (). It is similar to the incongruence of each chloroplast gene tree of available Bryophyte species, suggesting intensive researches to find genes causing incongruence with the classical phylogenetic relationship. More Bryophytes chloroplast genomes are required to resolve this incongruence precisely in the near future.
Figure 1. Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic tree of ten complete chloroplast genomes: Riccia fluitans (MK645896 in this study), Reboulia hemisphaerica (MK477551), Marchantia polymorpha subsp. ruderalis (NC_037507), Marchantia paleacea (NC_001319), Dumortiera hirsuta (NC_039590), Pellia endiviifolia (NC_019628), Aneura pinguis (NC_035617), Gymnomitrion concinnatum (NC_040133), Ptilidium pulcherrimum (NC_015402), and Sphagnum palustre (NC_030198) as an outgroup. Black bars in the right side indicate specific clades with labels. Phylogenetic tree was drawn based on neighbour joining tree from alignment of complete chloroplast genomes. The numbers above branches indicate bootstrap support values of neighbor joining and maximum likelihood phylogenetic trees from alignment of complete chloroplast genomes (black color) or 63 conserved genes (dart red color), respectively.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Ayub DM, Santos R, Costa D. 2014. Additions to the Ricciaceae flora of Rio Grande do Sul, including two remarkable records for the Brazilian liverwort flora. Phytotaxa. 161:294–300.
- Berrie GK. 1964. Experimental studies on polyploidy in liverworts I. The Riccia fluitans complex. Bryologist. 67:146–152.
- Cargill DC, Neal WC, Sharma I, Gueidan C. 2016. A preliminary molecular phylogeny of the genus Riccia L.(Ricciaceae) in Australia. Aust Syst Bot. 29:197–217.
- Carter AM. 1935. Riccia fluitans L.-A composite species. Bull Torrey Bot Club. 62:33–42.
- Crandall‐Stotler BJ, Hart ML, Long DG, Forrest LL. 2016. Divergence times and the evolution of morphological complexity in an early land plant lineage (Marchantiopsida) with a slow molecular rate. New Phytol. 209:1734–1746.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Kwon W, Kim Y, Park J. 2019a. The complete chloroplast genome of Korean Marchantia polymorpha subsp. ruderalis Bischl. & Boisselier: low genetic diversity between Korea and Japan. Mitochondr DNA B. 4:959–960.
- Kwon W, Kim Y, Park J. 2019b. The complete chloroplast genome sequence of Dumortiera hirsuta (Sw.) Nees (Marchantiophyta, Dumortieraceae). Mitochondr DNA B. 4:318–319.
- Kwon W, Kim Y, Park J. 2019c. The complete mitochondrial genome of Korean Marchantia polymorpha subsp. ruderalis Bischl. & Boisselier: inverted repeats on mitochondrial genome between Korean and Japanese isolates. Mitochondr DNA B. 4:769–770.
- Kwon W, Min J, Kim Y, Park J. 2019. The complete chloroplast genome sequence of Reboulia hemisphaerica (L.) Raddi (Aytoniaceae, Marchantiophyta). Mitochondr DNA B. 4:1459–1460.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.1
- Manju C, Rajesh K, Prakashkumar R. 2012. On the identity of Riccia fluitans (Ricciaceae: Marchantiophyta) in India. Acta Biol Plantarum Agriensis. 2:115–124.
- Park J, Kim Y, Xi H, Kwon W, Kwon M. Accepted. The complete chloroplast and mitochondrial genomes of Hyunsasi tree, Populus alba x Populus glandulosa (Salicaceae). DOI: 10.1080/23802359.2019.1598788
- Park J, Kwon W, Park J. 2019a. The complete mitochondrial genome of Camponotus concavus Kim & Kim, 1994 (Hymenoptera: Formicidae). Mitochondr DNA B. 4:1243–1244.
- Park J, Kwon W, Park J. 2019b. The complete mitochondrial genome of Siberian odorous ant, Dolichoderus sibiricus Emery, 1889 (Hymenoptera: Formicidae). Mitochondr DNA B. 4:525–526.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
