Abstract
Ormosia hosiei is a critically Endangered and important economic plant which is endemic in southern, eastern and central China. The complete chloroplast (cp) genome sequence of O. hosiei was determined by Next-generation sequencing technology. Total length of the cp genome is 172,210 bp, which contained a large single-copy (LSC) and a small single copy (SSC), separated by the inverted repeats (IR) of 40,377 bp. The cpDNA is structured with 144 genes, comprising 98 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. Phylogenetic analysis suggested that Ormosia is closely related to the genus Styphnolobium within the Fabaceae.
Ormosia hosiei, a nearly Threatened species and important economic plant, belongs to the family Fabaceae and is native to southern, eastern and central China. The number of natural populations of O. hosiei has declined rapidly and most of the extant populations are fragmental, due to habitat loss, over-harvesting, and poor reproduction capacity. The species is listed as the nearly Threatened level in the latest International Union for Conservation of Nature (IUCN) Red List (https://www.iucnredlist.org/species/32432/9706557). Furthermore, it is an important economic plant for its excellent wood for carving crafts and high-grade furniture. In the previous studies, some researches concerning O. hosiei focused on morphological and physiological characteristics and breeding techniques (Feng et al. Citation2007; Wang et al. Citation2015; Li et al. Citation2018) and the genetic diversity and population structures of O. hosiei distributed in Fujian, Hubei, and Zhejiang Province have been reported (Zhao et al. Citation2008; Zhang et al. Citation2012; Li et al. Citation2017). The complete cp genome sequence and accurate interpretation of O. hosiei distributed in Fujian Province was provided (Zhang et al. Citation2018). In this study, the complete cp genome of the wild-type O. hosiei distributed in Anhui Province was determined and described in order to provide basic genetic information about this species.
The fresh leaves of O. hosiei (Voucher number: HSP2018003) were collected from the Shexian County, Anhui Province, China. This ancient tree was transplanted from the south of China about 220 years ago. The cp genome sequence of O. hosiei was determined by Next-generation sequencing technology (Illumina Hiseq 2500, Shanghai Majorbio Bio-pharm Technology Co., Ltd. (SMBTC, Shanghai, China)). Genome sequences were picked out and assembled by the software SOAPdenovo v2.04 (Luo et al. Citation2012). The cp genome of O. hosiei (Genbank accession number MK450439) showed a typical structure with a length of 172,210 bp, which contained a large single-copy (LSC) and a small single copy (SSC) of 72,737 bp and 18,719 bp, respectively, separated by the inverted repeats (IR) of 40,377 bp. The cp DNA is structured with 144 genes, comprising 98 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. The overall base composition of the entire genome is as follows: A (32.0%), T (32.1%), C (18.2%) and G (17.7%), which the percentage of A + T is 64.1%.
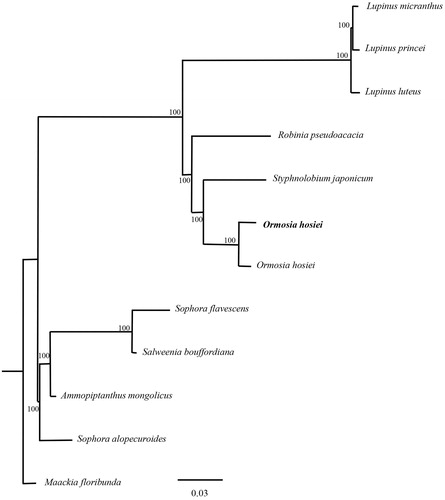
In order to validate the newly determined sequence, the whole cp genome sequence of the O. hosiei determined in this study and together with another sequence of O. hosiei and other 10 closely related species sequences from GeneBank to perform phylogenetic analysis. A maximum likelihood (ML) tree was constructed based on the dataset by the online tool RAxML (Kozlov et al. Citation2018). Phylogenetic analysis result suggested that Ormosia is closely related to the genus Styphnolobium within the Fabaceae () and was consistent with the previous research (Zhang et al. Citation2018).
Figure 1. The phylogenetic tree (ML) was constructed based on the dataset of whole chloroplast genome. Bold black branches highlighted the study species and corresponding phylogenetic classification. The analyzed species and corresponding NCBI accession number as follows: Lupinus micranthus (KU726828), Lupinus princei (KU726829), Lupinus luteus (KC695666), Robinia pseudoacacia (KJ468102), Styphnolobium japonicum (KY872756), Ormosia hosiei (MK450439), Ormosia hosiei (MG813874), Sophora flavescens (MH748034), Salweenia bouffordiana (MF449303), Ammopiptanthus mongolicus (KY034453), Sophora alopecuroides (MF156140), Maackia floribunda (KX388160).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Feng JG, Ji XL, Zhou ZC, Qu H, Huang HY, He YF. 2007. Cultivation techniques of Ormosia hosiei plantation. Chin Forest Sci Tech. 21:41–43.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2018. RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. BioRxiv 447110; doi:10.1101/447110
- Li FQ, Chen HW, Zhou ZC, Chu XL, Xu ZY, Xiao JJ. 2018. Analysis on variation of seed and seedling traits of superior trees of Ormosia hosiei and primary screening of optimum family. J. Plant Resour. & Environ. 27:57–65.
- Li FQ, Zhou ZC, Xie YJ. 2017. Genetic diversity and genetic differentiation of different populations of Ormosia hosiei in three watersheds. Mol Plant Breed. 15:4263–4274.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 1:18.
- Wang BS, He BT, Chen XL, Shen B, Hu GC. 2015. Seedling cultivation of Ormosia hosiei by root cutting. J Zhejiang Sci Technol. 35:65–68.
- Zhang JQ, Jiang MY, Zhu DD, Zhang JL, Zheng JZ, Deng CY. 2018. Characterization of the complete chloroplast genome of an endemic species of pea family in china, Ormosia hosiei (Fabaceae). Conserv Genet Resour. 5:1–4.
- Zhang R, Zhou ZC, Du KJ. 2012. Genetic diversity of natural populations of endangered Ormosia hosiei, endemic to China. Biochem Syst Ecol. 40:13–18.
- Zhao Y, He YF, Zhou ZC, Feng JG, Jin GQ, Wang BS. 2008. Genetic diversity of naturally reserved Ormosia hosiei populations in Zhejiang and Fujian Provinces. Chin. J. Ecol. 27:1279–1283.
