Abstract
Rosa lucidissima is a critically endangered wild rose species endemic to China with both ornamental value and medicinal usefulness. The complete chloroplast genome sequence of R. lucidissima was generated by de novo assembly using whole genome next-generation sequencing to provide genomic data for further conservation genetics of this species and for phylogenetic analysis of genus Rosa. The complete chloroplast genome was 156,588 bp in length, including two inverted repeat (IR) regions (26,048 bp), a small single-copy (SSC) region (18,765 bp), and a large single-copy (LSC) region (85,713 bp). The chloroplast genome encoded 129 genes, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The overall GC content of the whole genome was 37.25%. Phylogenetic analysis of 19 selected chloroplast genomes revealed that R. lucidissima was most closely related to R. chinensis var. spontanea.
Rosa lucidissima Lévl. (Rosaceae) is a wild rose species endemic to China. Very similar and close to R. chinensis var. spontanea, it is also thought to be one of the original species of section Chinenses (Ku Citation1985). With abundant beautiful purple red flowers and glabrous leathery leaves, R. lucidissima is an important wild rose germplasm resource. It is also a famous medicinal plant with the folk name of ‘Red Root’ to cure stomach trouble by the Zhuang Minorities in southeastern Yunnan and the Guangxi Zhuang Autonomous Region of China. It has been categorized as critically endangered (CR) with a status of ‘B1ab(iii)’ in the Threatened Species List of China’s Higher Plants (Qin et al. Citation2017). Here, we report the complete cp genome of R. lucidissima, to provide genomic data for further conservation genetics of this species and for the phylogenetic analysis of genus Rosa.
Fresh leaves of R. lucidissima were collected from Malipo County (23°16'11″N, 104°30'37″E, 1573 m) in southeastern Yunnan. The voucher specimen (JHYws001) was deposited in the Herbarium of Southwest Forestry University. Total genomic DNA was extracted using the modified CTAB method (Yang et al. Citation2014). A shotgun library was prepared and sequenced using the IlluminaHiseq 2000 (Illumina, San Diego, CA, USA) at Novogene (Beijing, China). All raw reads (about 3.5 Gb) were filtered using NGSQCToolkit_v2.3.3 with default parameters to obtain clean reads (Patel and Jain Citation2012). The genome was de novo assembled by the GetOrganellepipleline (https://github.com/Kiggerm/GetOrganell), automatically annotated by the CpGAVAS pipeline (Liu et al. Citation2012), and then adjusted by Geneious 8.1 (Kearse et al. Citation2012). To determine the phylogenetic position of R. lucidissima, its LSC, SSC, one IR region sequence, and those of other 15 published chloroplast genomes of Rose family were used to construct a maximum likelihood (ML) tree. The sequences were aligned by the MAFFT (Katoh and Standley Citation2013) and the tree was constructed by RAxML 8.2.11 (Stamatakis Citation2014) with the nucleotide substitution model of GTR + G and node support estimated by means of bootstrap analysis with 1000 replicates.
The plastome of R. lucidissima (GenBank accession number: MK782979) was a typical quadripartite circular with 156,588bp in length, comprising a large single-copy (LSC) region of 85,713 bp and a small single-copy (SSC) region of 18,765 bp, separated by two inverted repeat (IR) regions of 26,048bp, respectively. The GC content was 37.25% overall. The IR regions had a higher GC content (42.74%) than LSC (35.20%) and SSC (31.22%) regions. The plastome of R. lucidissima included 57.79% coding sequences (50.22% protein-coding regions, 1.8% tRNAs, and 5.78% rRNAs) and 42.21% non-coding sequences (13.47% introns and 28.73% intergenic spacers). The plastome encoded 129 genes, including 84 protein-coding genes (PCGs), 37 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. The trnK-UUU gene has the largest intron of 2498 bp, in which the matK gene was located.
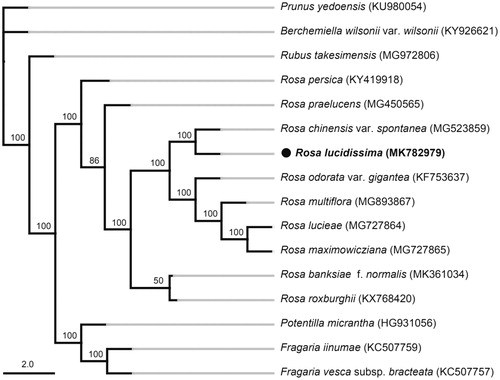
The phylogenetic tree showed that species from genus Rosa formed a monophyletic clade with 100% bootstrap value (), in which R. lucidissima was mostly closely related to R. chinensis var. spontanea.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Ku TC. 1985. Rosa. In: Editorial Board of the Flora Republicae Popularis Sinicae, editors. Flora reipublicae popularis sinicae.Volume 37. Beijing: Science Press; p. 360–455.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619.
- Qin H, Yang Y, Dong S, He Q, Jia Y, Zhao L, Yu S, Liu H, Liu B, Yan Y, et al. 2017. Threatened species list of China’s higher plants. Biodivers Sci. 25:696–744.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031.