Abstract
Semenovia gyirongensis is an endemic species in China. The complete chloroplast (cp) genome sequence of S. gyirongensis was generated by de novo assembly using whole genome next-generation sequencing. The cp genome was a circular structure and 147,246 bp in length, composed of four distinct regions such as large single-copy (LSC) region of 92,833 bp, small single-copy (SSC) region 17,545 bp, and a pair of inverted repeat regions of 36,868 bp. The cp genome contained 129 genes, including 85 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The GC content was 37.6%. Phylogenetic analysis of 21 representative plastomes indicated that the S. gyirongensis was close to Pastinaca pimpinellifolia, Cnidium officinale, and Ligusticum sinense in the family Apiaceae.
Semenovia gyirongensis Q.Y.Xiao & X.J.He (Apiaceae, Apioideae) is a perennial herb, which is endemic to Gyirong County, Xizang, China, growing on screes, rocky slopes and sandy places, at elevations between 4000 and 4150 m (Xiao et al. Citation2017). In field investigation, we found that the taxon is locally common and used for excessive grazing use. Because of its localized distribution and grazing pressure, it was “Vulnerable” (VU) (Xiao et al. Citation2017). Therefore, this species should be urgently scientifically and effectively conserved. Here, we reported the complete chloroplast (cp) genome of S. gyirongensis for phylogenetic studies and the protection of genetic resources.
The mature leaves of S. gyirongensis were obtained from a rocky slope near Longda (28°45'06''N, 85°18'13''E, altitude 4023 m), Woma village, Gyirong County, Xizang, China, and preserved in liquid nitrogen for further study. Voucher specimens (voucher number: xqy-20160730-01) were deposited in the herbarium of Natural History Museum of Sichuan University (SZ). Total genomic DNA of S. gyirongensis was isolated using the Plant Genomic DNA Kit (TIANGEN Biotech., Beijing, China) and sequenced on an Illumina HiSeq × Ten platform (Illumina, San Diego, CA, USA). Approximately 5 Gb of raw data were generated through pair-end 150 bp sequencing. Adapters and low-quality reads were removed and high-quality reads were used for the cp genome assembly using SOAPdenovo2 (Luo et al. Citation2012). The resulting contigs were linked based on overlapping regions after being aligned to Cnidium officinale (NC_039760) using Geneious version 11.0.4 (Kearse et al. Citation2012). The plastome was annotated using Geneious version 11.0.4 against the plastome of Cnidium officinale (NC_039760), coupled with manual check and adjustment.
The circular double-stranded complete chloroplast genome (GenBank Accession number: MK757488) was 147,246 bp in length, exhibiting a quadripartite structure with a large single-copy region (LSC) of 92,833 bp and a small single-copy region (SSC) of 17,545 bp joined by two identical inverted repeat regions (IRa and IRb, 18,434 bp each). The overall GC content was 36.7% and the plastome contained 129 genes, including 85 protein-coding genes, 8 rRNA genes, and 36 tRNA genes were annotated. A total of 15 genes duplicated in the inverted repeat regions including six tRNA, four rRNA, and five protein-coding genes.
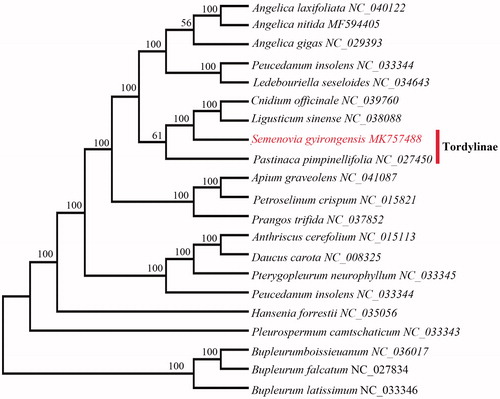
To confirm the phylogenetic position of S. gyirongensis, 21 representative species of allied genera were aligned using MAFFT v.7 (Katoh and Standley Citation2013) and maximum likelihood (ML) analysis was conducted using RAxML (Stamatakis Citation2014) with 1000 bootstraps under the GTRGAMMAI substitution model. The phylogenetic tree () indicated that S. gyirongensis was closely related to Pastinaca pimpinellifolia, Cnidium officinale, and Ligusticum sinense. This published S. gyirongensis chloroplast genome will provide useful information for phylogenetic studies and conservation genetics.
Acknowledgements
The authors thanks Dr. Deng-Feng Xie and Ma. Lu Kang for their help in preparing this paper.
Disclosure statement
No potential conflict of interest was reported by the authors.
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Funding
References
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Pan Q, Liu YJ, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Xiao QY, Tan JB, Hu HY, Zhou SD, He XJ. 2017. Semenovia gyirongensis (Apiaceae), a new species from Xizang, China. Phytokeys. 82:57–72.