Abstract
Malania oleifera is a rare and threatened medicinal plant in China. In this study, the complete chloroplast genome (cp) of M. oleifera was sequenced, assembled, and analyzed. The circular complete cp genome of M. oleifera was 158,163 bp in length and contained a large single copy region (LSC, 87,212 bp), a small single copy region (SSC, 439 bp), and 2 short inverted repeat (IRa and IRb) regions (35,256 bp). The cp genome encodes 97 unique genes, including 62 protein-coding genes, 30 transfer RNA genes, 4 ribosomal RNA genes, and a pseudogene. Phylogenetic analysis of M. oleifera showed that M. oleifera was the sister group with Champereia manillana from Opiliaceae and Osyris alba from Santalaceae.
Malania oleifera Chun et Lee, an evergreen broad-leaved wood plant endemic to south-western China, had original properties on dissection and relatively evolved characteristics. The oil in the seeds can be used as ideal raw materials of muscone (Liang et al. Citation2003). Now, M. oleifera is rated as the national second protection species and is becoming endangered in China due to habitat loss. On the other hand, the origin of the taxa is not accurately resolved and taxonomical position is still unclear. Hence, a better understanding of this species has a great potential in species discrimination and protection of species. Hence, a better understanding of this species has a great potential in species discrimination and protection of species. In this study, we characterized the complete chloroplast (cp) genome sequence of M. oleifera based on the genome skimming sequencing data, detected the occurrence, type, and constructed the phylogenetic tree based on the neighbour-joining (NJ) method.
The fresh leaves of M. oleifera were collected from the Arboretum of the Chinese Academy of Forestry. The voucher specimen was deposited at Herbarium, Kunming Institute of Botany, CAS (KUN). Total genomic DNA was isolated from fresh leaves using the modified cetyltrimethyl ammonium bromide (CTAB) method (Doyle Citation1987) to construct chloroplast DNA libraries. The extracted DNA was sequenced using the Illumina Hiseq2000 (Illumina, CA, USA). In all, 186 M of 150 bp raw paired reads were retrieved. Prior to chloroplast de novo assembly, low-quality reads were filtered out, and resultant clean reads were assembled using GetOrganelle pipeline (https://github.com/Kinggerm/GetOrganelle). All the contigs were checked against the reference genome of Champereia manillana (KY436366), using BLAST (https://blast.ncbi.nlm.nih.gov/) and aligned contigs were oriented according to the reference genome.
The genome was automatically annotated by using the CpGAVAS pipeline (Liu et al. Citation2012) and start/stop codons and intron/exon boundaries were adjusted in Geneious 8.1 (Kearse et al. Citation2012). The tRNA was identified through tRNAscan-SE v2.0 (Lowe and Chan Citation2016). Sequence data was deposited into GenBank. A physical map of the cp genome was generated using the online tool OGDraw v1.2 (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. Citation2007). The annotated cp genome sequence was submitted to the GenBank under Accession Number of MK764537.
The circular complete cp genome of M. oleifera was 158,163 bp in length. It had the typical quadripartite structure and contained a large single copy region (LSC, 87,212 bp), a small single copy region (SSC, 439 bp) and two short inverted repeat (IRa and IRb) regions (35,256 bp). We found the length of SSC of this species smaller than other closely related genetic species, such as Erythropalum scandens (18,567 bp) and Osyris alba (13,973 bp).
The encoded genome sequence of M. oleifera contained 97 unique genes, including 62 genes, 4 rRNA, 30 tRNA, and a pseudogene (ΨndhB). Among these annotated genes, most of genes occurred as single-copy, while 11 PCGs (ccsA, psaC, rpl2, rpl23, rpl32, rps12, rps15, rps19, rps7, ycf1, ycf2), 8 tRNAs (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnL-UAG, trnN-GUU, trnR-ACG, trnV-GAC), 4 rRNA genes (rrn16, rrn23, rrn4.5, rrn5), and 1 pseudogene had two copies. In addition, two PCGs (clpP and ycf3) had two introns each; eight PCGs (atpF, petB, petD, rpl2, rpl16, rpoC1, rps12 and rps16) contained one intron, and six tRNAs (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA and trnV-UAC) contained one intron. The overall GC content of the cp genome was 36.7%, while that of LSC, SSC, and IR regions was 34.4, 25.7, and 39.6%, respectively. We found SSC region has relatively low GC ratio than other genetically close species such as E. scandens (32.3%).
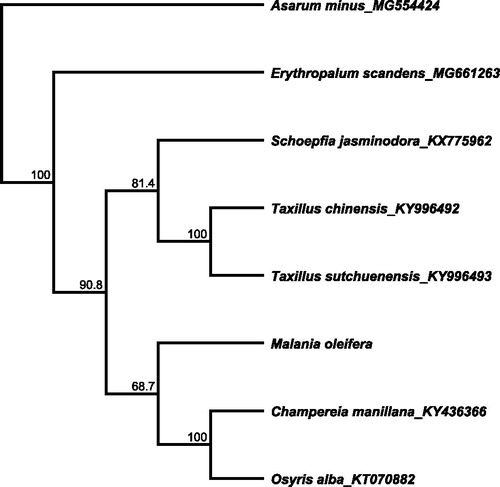
A total of eight cp genome sequences were selected to infer the phylogenetic relationships among the main representative species of Rhoeadales and related orders with Asarum minus (Aristolochiaceae) as the outgroup. The combined datasets based on plastid genomes of 8 species were aligned by MAFFT v7.307 (Katoh and Standley Citation2013). A neighbour-joining (NJ) phylogenetic tree was constructed using Geneious 8.1 (Kearse et al. Citation2012) with the Tamura–Nei genetic distance model and a total of 1000 bootstrap replicates were performed. The topology of the phylogenetic tree showed that the species of M. oleifera was the sister group with C. manillana from Opiliaceae and O. alba from Santalaceae (). The complete cp genome information reported in this study will be a valuable resource for future studies on conservation genetics, taxonomy, phylogeny, and breeding of the Olacaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647.
- Liang YF, Shu-Guang WU, Xiang-Dong LI. 2003. Study on the endangered causes for Malania oleifera. Guihaia. 23:404–407.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52:267–274.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:54–57.