Abstract
In this study, the complete mitogenome of brownbranded moon jellyfish Aurelia limbata (Cnidaria, Semaeostomeae, Ulmaridae) was sequenced and analyzed. The linear mitogenome is 16,953 bp long with 37.3% A, 16.8% C, 15.5% G, and 30.4% T nucleotide distributions. Total AT% content is 67.7%, which is the highest AT% content among available Aurelia species complete mitogenome records. Furthermore, phylogenetic relationships of A. limbata in the family Ulmaridae were investigated. According to protein-coding genes based phylogenetic tree, the A. aurita species collected from Korea water is the closest related species to A. limbata.
Aurelia is a commonly distributed scyphozoan genus, which mostly known as moon jellyfish. They are important research organisms for studies in a wide range, such as protein chemistry, developmental biology, ecology, and hydrodynamics (Arai Citation1997). Although they are well-studied model organisms, their taxonomy is still unclear because of the lack of molecular data (Dong et al. Citation2015). In the present study, we reported the first complete mitogenome from brownbranded moon jellyfish Aurelia limbata and investigated the molecular phylogeny of the genus Aurelia.
The specimens were collected in Namae port (37°56′43.0″N 128°47′15.5″E), Yangyang-gun, Korea, on 5 January 2016. Total genomic DNA was extracted from the jellyfish umbrella using the cetyltrimethylammonium bromide method (Ausubel et al. Citation1989) and the remained part of the specimen was stored in Department of Biotechnology, Sangmyung University (Accession no: S002). The mitochondrial DNA was sequenced by NGS technology with Miseq system (Illumina, CA, USA). After sequencing, MITObim (Hahn et al. Citation2013) and MITOS web server (Bernt et al. Citation2013) were used for assembly and annotation of the paired-end reads, respectively. Furthermore, phylogenetic relationships of the A. limbata within the family Ulmaridae were investigated by reconstruction of the phylogenetic tree predicted with the software MEGA 7.0 (Kumar et al. Citation2016). For reconstruction of the phylogenetic tree, concatenated amino acid sequences of 13 mitochondrial protein-coding genes were used.
The complete mitogenome of A. limbata (GenBank Accession no: MK527107) is 16,953 bp in length. The structure and gene orientation of the mitogenome is identical with the other Aurelia species, such as A. aurita (HQ694729 and DQ787873), Aurelia sp. 3 (LC005413), and Aurelia sp. 4 (LC005414). It consists of 13 protein-coding genes, 2 tRNAs, and 2 rRNAs. Among 17 genes, only cox1 and 16S rRNA encoded in the minority strand the rest 15 genes encoded in the majority strand. In addition to the typical set of protein-coding genes, two unknown open-reading frames (ORFs) have been found, which were located between 16S rRNA and cytb. These ORFs are also common for Aurelia mitogenome (Shao et al. Citation2006; Park et al. Citation2012).
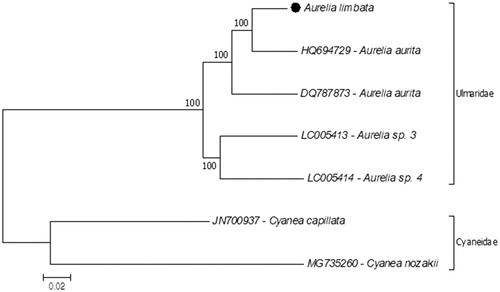
According to protein-coding genes-based phylogenetic tree, A. aurita (HQ694729) is the closest related species to A. limbata. These two species were collected from Korea. They are clustered together with another A. aurita (DQ787873) record obtained from the White Sea, Russia (). These results show similarity with the previous molecular phylogenetic study based on nuclear internal transcribed spacer one (ITS-1) and mitochondrial cytochrome oxidase c subunit I (COI) of Aurelia species (Dawson and Jacobs Citation2001). The same study also described the possibility of cryptic species in A. aurita (Dawson and Jacobs Citation2001). This could be the reason for the relationships of A. limbata and two A. aurita in the phylogenetic tree. Although Aurelia is the most-studied genus of all scyphozoans, its taxonomy and evolutions are still poorly known. For better resolution, more complete mitogenome data should be recorded from the family.
Figure 1. Molecular phylogeny of Aurelia limbata in the family Ulmaridae. Two species from the family Cyaneidae represent outgroup. The phylogeny tree reconstructed according to protein-coding genes of the mitochondrial genome with a maximum likelihood statistical method by using MEGA 7.0 software. mtREV with JTT matrix-based model used for amino acid substitution and bootstrap method replicated 1000 times for the test of phylogeny. The complete mitochondrial genomes were retrieved from the GenBank. Aurelia limbata highlighted with a black dot.
Acknowledgement
We thank Dr. Jinho Chae for jellyfish samplings.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Arai MN. 1997. A functional biology of Scyphozoa. London: Chapman & Hall.
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1989. Current protocols in molecular biology. New York: John Wiley and Sons.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenetics Evol. 69:313–319.
- Dawson MN, Jacobs DK. 2001. Molecular evidence for cryptic species of Aurelia aurita (Cnidaria, Scyphozoa). Biol Bull. 200:92–96.
- Dong Z, Liu Z, Liu D. 2015. Genetic characterization of the scyphozoan jellyfish Aurelia spp. in Chinese coastal waters using mitochondrial markers. Mol Syst Ecol. 60:15–23.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:129.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Park E, Hwang DS, Lee JS, Song JI, Seo TK, Won YJ. 2012. Estimation of divergence times in cnidarian evolution based on mitochondrial protein-coding genes and the fossil record. Mol Phylogenetics Evol. 62:329–345.
- Shao Z, Graf S, Chaga OY, Lavrov DV. 2006. Mitochondrial genome of the moon jelly Aurelia aurita (Cnidaria, Scyphozoa): a linear DNA molecule encoding a putative DNA-dependent DNA polymerase. Gene. 381:92–101.
