Abstract
It is the first report on complete chloroplast genome of Alsophila costularis, a least concerned relict tree fern. The genome size is 156,675 bp, with the typical quadripartite structure containing a pair of inverted repeats (IRs) of 24,356 bp separated by a large single-copy (LSC) region and a small single-copy (SSC) region of 86,338 bp and 21,625 bp, respectively. A total of 133 genes are predicted, including 89 protein-coding genes, eight ribosomal RNA genes, 33 tRNA genes, and three pseudogenes. The ML phylogenetic tree revealed A. costularisis as sister to Alsophila spinulosa.
Alsophila costularis Baker, known as Cyathea chinensis Copel, is a relict fern with the accepted name in Cyatheaceae. Its trunk is 1.8–7.5 m tall with loose crown and rhizomes are covered with densely and scaly dark brown scales. Fronds are bipinnate and 1–2 m long. As a tree fern native to Yunnan in China, it occurs in India, Nepal, Myanmar, Laos, and Vietnam as well. The species grows in ravine forests at an altitude of 700–2100 m (Zhang and Nishida Citation2013). Different from other ‘living fossil’ ferns in Cyatheaceae, A. costularis is least concerned. The current report focuses only on tissue culture (Cheng and Liu Citation1992), gametophyte development (Wang et al. Citation2007), and genetic diversity (Li et al. Citation2010). Due to a second-class protected plant, considerable attention should be paid to the protection of A. costularis based on chloroplast genetic variation. In addition, A. costularis is very critical for the classification and phylogeny of Cyatheaceae (Ching Citation1978; Wang et al. Citation2003). Hence, it is necessary to acquire its whole chloroplast (cp) genome sequence for conservation genetics and the chloroplast phylogenomics of Cyatheaceae.
Fairy Lake Botanical Garden, Shenzhen, Chinese Academy of Sciences (22°34′45.69″N, 114°10′7.92″E) provided plant material of A. costularis, which is stored in the Herbarium of Sun Yat-sen University (SYS; voucher: SS Liu 201619). Tiangen Plant Genomic DNA Kit (Tiangen Biotech Co., Beijing, China) was used to extract total genomic DNA from leaves, which was further broken and used for Illumina library preparation. Genome was sequenced on a Hiseq 2500 platform (Illumina Inc., San Diego, CA) with paired-end reads of 300 bp. We obtained 12,053,058 raw reads. After filtering out and trimming with Trimmomatic v0.32 (Bolger et al. Citation2014), the remaining 11,109,827 high-quality clean data with an average coverage of 100× were used to de novo assemble the chloroplast genome in Velvet (Zerbino and Birney Citation2008) by mapping to referenced-based chloroplast genomes of A. spinulosa (NC_012818). Using the same reference, all genes were annotated with the Dual Organellar GenoMe Annotator (DOGMA; Wyman et al. Citation2004) and tRNAscan-SE programs (Lowe and Eddy Citation1997). We used RAxML v8.0 (Stamatakis Citation2014) with 1000 bootstraps to reconstruct a maximum-likelihood (ML) phylogeny of 13 ferns including Diplopterygium glaucum as outgroup.
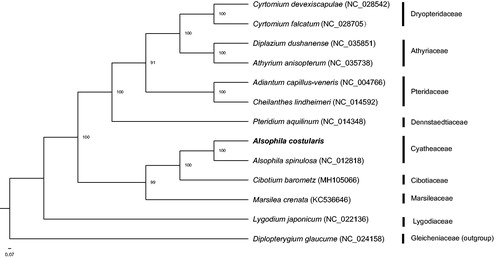
Alsophila costularis has a total length 156,675 bp with the typical quadripartite structure including a pair of inverted repeats (IRs) of 24,356 bp separated by a large single-copy (LSC) region and a small single-copy (SSC) region of 86,338 bp and 21,625 bp, respectively (Genbank accession number: MH684489). The genome contains 133 genes, including 89 protein-coding genes, eight ribosomal RNA genes, 33 tRNA genes, and three pseudogenes. Among them, 15 genes contain one intron, while three genes have two introns. Thirteen genes are duplicated, including four protein-coding genes, five tRNA genes, and four rRNA genes. The overall A/T content was 60%, while the corresponding values of the LSC, SSC and IR regions were 60%, 62% and 57%, respectively. The phylogenetic analysis revealed A. costularis as sister to A. spinulosa with 100% bootstrap support (). The complete chloroplast genome of A. costularis lays a solid foundation for further phylogenetic studies.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Cheng ZY, Liu DH. 1992. Tissue culture of Alsophila costularis. Plant Physiol Commun. 28:210–211.
- Ching RC. 1978. The Chinese fern families and genera: systematic arrangement and historical origin. Acta Phytotaxon Sin. 16:1–19. (in Chinese)
- Li Y, Hou KL, Ying ZM, Song GW. 2010. ISSR analysis of the genetic diversity within two populations of relict plant, Alsophila costularis (Cyatheaceae). Genomics Appl Biol. 29:679–684. (in Chinese)
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wang JJ, Zhang XC, Liu BD, Cheng X. 2007. Gametophyte development of three species in Cyatheaceae. J Trop Subtrop Bot. 15:115–120.
- Wang T, Su YJ, Zheng B, Li XY, Chen GP, Zeng QL. 2003. Phylogenetic analysis of the chloroplast trnL intron and trnL-trnF intergenic spacer sequences of the Cyatheaceae plants from China. J Trop Subtrop Bot. 11:137–142. (in Chinese)
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhang XC, Nishida H. 2013. Cyatheaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 2–3 (Pteridophytes). Beijing: Science Press; p. 134–138.