Abstract
Genomic analysis of the marine alga Corallina chilensis from Tomales Bay, California, USA, resulted in the assembly of its complete mitogenome (GenBank accession number MK598844) and plastid genome (GenBank MK598845). The mitogenome is 25,895 bp in length and contains 50 genes. The plastid genome is 178,350 bp and contains 233 genes. The organellar genomes share a high-level of gene synteny to other Corallinales. Comparison of rbcL and cox1 gene sequences of C. chilensis from Tomales Bay reveals it is identical to three specimens from British Columbia, Canada and very similar to a specimen of C. chilensis from southern California. These genetic data confirm that C. chilensis is distributed in Pacific North America.
Corallina chilensis Decaisne (Harvey Citation1849) is a calcified, intertidal marine red alga originally collected by Charles Darwin from Valparaíso, Chile. Harvey’s species diagnosis described it as being 2.5–5 cm in height with slender stems below and broader and bipinnate stems above (Harvey Citation1849). The binomial was long regarded as C. officinalis var. chilensis (Decaisne) Kützing (Citation1858), but was recently elevated to specific status by Williamson et al. (Citation2016). We performed high-throughput sequencing on a specimen of C. chilensis from Tomales Bay, California, to determine its genomic structure and genetic relationship to other species of Corallina.
DNA was extracted from C. chilensis (Specimen Voucher – UC2050474) following the protocol of Lindstrom et al. (Citation2011). The 150 bp PE Illumina library construction and sequencing was performed using myGenomics, LLC (Alpharetta, Georgia, USA). The genomes were assembled using default de novo settings in Geneious Prime (Biomatters Limited, Auckland, New Zealand) and annotated using Geneious Prime and blastx, NCBI ORFfinder, and tRNAscan-SE 1.21 (Schattner et al. Citation2005). The C. chilensis mitogenome was aligned to other mitogenomes with MAFFT (Katoh and Standley Citation2013). The phylogenetic analysis was executed using RAxML-NG (Kozlov et al. Citation2018) with the GTR + gamma model and 100 bootstraps. The tree was visualized with TreeDyn 198.3 at Phylogeny.fr (Dereeper et al. Citation2008) and pairwise distances were calculated using default parameters in DIVEIN with the GTR model (Deng et al. Citation2010).
The mitogenome of C. chilensis is 25,895 bp in length and contains 50 genes. It is A + T rich (70.5%) and contains 23 tRNA (trnG, trnL, trnM, trnR, trnS are duplicated), 5 ribosomal proteins (rpl16, rpl20, rps3, rps11, rps12), 2 rRNA (rrl, rrs), orf158, TatC, ymf39, and 17 other genes involved in electron transport and oxidative phosphorylation. The plastid genome of C. chilensis is 178,350 bp and contains 233 genes. The genome is also A + T biased (70.0%) and contains 47 ribosomal proteins, 31 tRNA, 30 photosystem I and II, 28 ycf, 10 phycobiliprotein, 8 cytochrome b/f complex, 8 ATP synthase, 4 RNA polymerase, 4 orfs, 3 rRNA, and 60 other genes. The mitogenome and plastid genome of C. chilensis are similar in length, content, and organization to other coralline red algae (Janouškovec et al. Citation2013; Bi et al. Citation2016; Williamson et al. Citation2016; Lee et al. Citation2018; Gabrielson et al. Citation2018; Bustamante et al. Citation2019).
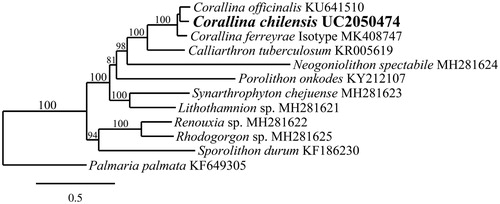
Phylogenetic analysis of the C. chilensis mitogenome resolved it in a fully supported clade with C. officinalis L. and C. ferreyrae E.Y. Dawson, Acleto & Foldvik (). The mitogenome of C. chilensis differed in pairwise distance from C. officinalis by 9.8% and C. ferreyrae by 10.8%. The plastid genome of C. chilensis differed from C. ferreyrae by 2.9%. A BLAST analysis of cox1 and rbcL sequences of C. chilensis from Tomales Bay identified three identical sequences from British Columbia, Canada (as C. frondescens) (Hind and Saunders Citation2013) and a nearly identical sequence from a specimen of C. chilensis from Monarch Bay, Orange County, California (Williamson et al. Citation2015).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bi G, Liu G, Zhao E, Du Q. 2016. Complete mitochondrial genome of a red calcified alga Calliarthron tuberculosum (Corallinales). Mitochondrial DNA. 27:2554–2556.
- Bustamante DE, Calderon MS, Hughey JR. 2019. Conspecificity of the Peruvian Corallina ferreyrae with C. caespitosa (Corallinaceae, Rhodophyta) inferred from genomic analysis of the type specimen. Mitochondrial DNA Part B. 4:1285–1286.
- Deng W, Maust BS, Nickle DC, Learn GH, Liu Y, Heath L, Kosakovsky Pond SL, Mullins JI. 2010. DIVEIN: a web server to analyze phylogenies, sequence divergence, diversity, and informative sites. Biotechniques. 48:405–408.
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. 2008. Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469.
- Gabrielson PW, Hughey JR, Diaz-Pulido G. 2018. Genomics reveals abundant speciation in the coral reef building alga Porolithon onkodes (Corallinales, Rhodophyta). J Phycol. 54:429–434.
- Harvey WH. 1849. Nereis australis, or algae of the southern ocean: being figures and descriptions of marine plants, collected on the shores of the Cape of Good Hope, the extra-tropical Australian colonies, Tasmania, New Zealand, and the Antarctic regions; deposited in the Herbarium of the Dublin University. Part 2. London: Reeve Brothers.
- Hind KR, Saunders GW. 2013. A molecular phylogenetic study of the tribe Corallineae (Corallinales, Rhodophyta) with an assessment of genus-level taxonomic features and descriptions of novel genera. J Phycol. 49:103–114.
- Janouškovec J, Liu SL, Martone PT, Carré W, Leblanc C, Collén J, Keeling PJ. 2013. Evolution of red algal plastid genomes: ancient architectures, introns, horizontal gene transfer, and taxonomic utility of plastid markers. PLoS One. 8:e59001
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2018. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. bioRxiv 447110; doi: https://doi.org/10.1101/447110.
- Kützing FT. 1858. Tabulae phycologicae; oder, Abbildungen der Tange. Vol. VIII pp. i-ii, 1-48, 100 plates. Nordhausen: Gedruckt auf kosten des Verfassers (in commission Bei W. Köhne).
- Lee JM, Song HJ, Park SI, Lee YM, Jeong SY, Cho TO, Kim JH, Choi HG, Choi CG, Nelson WA, et al. 2018. Mitochondrial and plastid genomes from coralline red algae provide insights into the incongruent evolutionary histories of organelles. Genome Biol Evol. 10:2961–2972.
- Lindstrom SC, Hughey JR, Martone PT. 2011. New, resurrected and redefined species of Mastocarpus (Phyllophoraceae, Rhodophyta) from the northeast Pacific. Phycologia. 50:661–683.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucl Acids Res. 33:686–689.
- Williamson CJ, Walker RH, Robba L, Yesson C, Russell S, Irvine LM, Brodie J. 2015. Toward resolution of species diversity and distribution in the calcified red algal genera Corallina and Ellisolandia (Corallinales, Rhodophyta). Phycologia. 54:2–11.
- Williamson C, Yesson C, Briscoe AG, Brodie J. 2016. Complete mitochondrial genome of the geniculate calcified red alga, Corallina officinalis (Corallinales, Rhodophyta). Mitochondrial DNA Part B. 1:326–327.