Abstract
In this study, the complete mitochondrial genome of Ilex pubescens was recovered through Illumina sequencing data. This complete mitochondrial genome of I. pubescens was 517,520 bp in length and contained a sparse gene coding structure, which account for 12.6% of the whole genome. Mt genome of I. pubescens encoded 54 genes including 33 protein-coding genes, 18 tRNA genes and 3 ribosomal RNA genes. The overall GC content of I. pubescens mt genome is 45.6%. By phylogenetic analysis using whole genome-wide alignments through IQ-TREE, I. pubescens existed in a unique branch as a distinct order, which showed a relatively close relationship within Campanulids clade. This complete mitochondrial genome can be subsequently used for further evolution studies of asterids plants.
Ilex pubescens is a Chinese herbal medicine widely distributed in south China. Its root has been well-known for its medicinal use in treating cardiocerebral, hypercholestaemia, and arterial thrombosis (Zhang et al. Citation2010). In this study, the complete mitochondrial genome of I. pubescens was recovered through Illumina sequencing data. This complete mitochondrial genome can be subsequently used for evolution studies of asterids plants.
Fresh leaves of I. pubescens were supplied by medical plants garden in Guiyang University of Traditional Chinese Medicine. Whole genomic DNA was extracted using CTAB method. Isolated DNA was used in High-through sequencing on Illumina Hiseq 2000 platform. Both dried leaves and isolated DNA was stored at −20 °C in College of Life Science of Northwest A&F University. The raw reads sequenced by Illumina Hiseq 2000 platform were trimmed after quality filtration, then clean reads were assembled by SPAdes version 3.11.0 (Bankevich et al. Citation2012) based on default settings. We used other mitochondrial genomes from Campanulids as reference sequences to align the contigs and identify gaps. To fill the gap, Price (Ruby et al. Citation2013) and MITObim version 1.8 (Hahn et al. Citation2013) were applied and Bandage (Wick et al. Citation2015) was used to verify the circular structure. The mean sequencing coverage of this mt genome is 76X. This complete sequence was primarily annotated by Geseq (Michael et al. Citation2017) combined with manual correction. All tRNAs were confirmed using the tRNAscan-SE search server (Lowe and Eddy Citation1997). Other protein-coding genes were verified by BLAST search on the NCBI website (http://blast.ncbi.nlm.nih.gov/), and manual correction for start and stop codons were conducted. This complete mt genome sequence together with gene annotations was submitted to GenBank under the accession numbers of MK714017.
The mitochondrial genome of I. pubescens, with a length of 517,520 bp and contained a sparse gene coding structure, which account for 12.6% of the whole genome. This mt genome possesses 54 genes, including 33 protein-coding genes, 3 ribosomal RNA genes (26S, 18S, and 5S ribosomal RNA), and 18 tRNA genes. The overall GC content of the mt genome is 45.6%. The genome structure, gene order, GC content are unique when compared to other mitochondrial genomes in Campanulids.
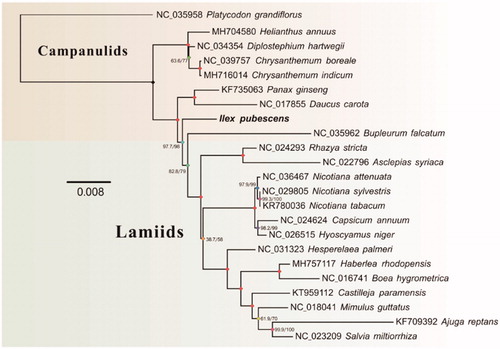
For phylogenetic analysis assessing the relationship of this species, we selected other 22 mitogenomes from asterids to construct whole genome-wide alignments using HomBlocks (Bi et al. Citation2018), resulting in 26,784 characters in total, including almost all whole or partial PCGs and rRNA genes. The phylogeny tree was constructed by IQ-TREE version 1.6.6 (Nguyen et al. Citation2015) according to the optimal substitution models under the rapid bootstrap algorithm (1000 replicates) (). As expected, I. pubescens existed in a unique branch as a distinct order, which showed a relatively close relationship within the clade of Campanulids.
Acknowledgments
The authors are grateful to the anonymous reviewers for their critical comments.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bankevich A, Sergey N, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129–e129.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Michael T, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Nguyen LT, Schmidt HA, Haeseler A, von Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Ruby JG, Bellare P, DeRisi JL. 2013. PRICE: software for the targeted assembly of components of (Meta) genomic sequence data. G3 (Bethesda). 3:865–880.
- Wick RR, Schultz MB, Schultz J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31:3350–3352.
- Zhang CX, Lin CZ, Xiong TQ, Zhu CC, Yang JY, Zhao ZX. 2010. New triterpene saponins from the root of Ilex pubescens. Fitoterapia. 81:788–792.