Abstract
The complete chloroplast genome of Allium macrostemon Bunge was determined. The length of the complete chloroplast genome is 153,158 bp. The whole chloroplast genome includes four regions: 82,700 bp long single-copy (LSC) and 17,600 bp small single-copy (SSC) regions separated by a pair of 26,429 bp inverted repeat (IR) regions. The overall GC content of the chloroplast genome is 36.7% and those in the LSC, SSC, and IR regions are 34.6, 29.1, and 42.7%, respectively.
Allium macrostemon, a species of wild Allium plant that naturally distributed in East Asia, grows wildly in hills, slopes, valleys, and plains at an elevation of 0–1600 m, even 3000 m. Bulbs of A. macrostemon are usually using as traditional Chinese medicine in China (Li et al. Citation2014), and it can also be eaten as a wild vegetable. Chloroplast genomes had been widely used in phylogenetic analysis and molecular identification of plant species in recent years (Moore et al. Citation2010; Kuang et al. Citation2011), although several phylogenetic studies reported nuclear and chloroplast sequences of A. macrostemon, the complete chloroplast genome sequence is not available till now. In this study, we get the complete chloroplast genome of A. macrostemon for the following research.
The leaves of A. macrostemon were collected in China (Mountain Baohua, Jiangsu, China), the specimens were deposited in Sichuan University Herbarium (SZ) and the voucher number is XFM20180817-2. The total DNA was extracted from dry leaves using Plant Genomic DNA Kit. The complete chloroplast genome was sequenced using an Illumina HiSeq 2500 system at Novogene (Beijing, China), and the chloroplast was completed using NOVOPlasty (Dierckxsens et al. Citation2017), the seed sequence is Allium cepa (KM088014). The chloroplast genome of A. macrostemon was annotated using Geneious version 10.2 (Kearse et al. Citation2012) based on published chloroplast genome of A. cepa (KM088014). Then, the annotated chloroplast genome was deposited into the GenBank, with the accession number MK751472. The phylogenetic relationship between A. macrostemon and related species was performed using RAxML-HPC BlackBox version 8.1.24 software (Stamatakis Citation2006) with the GTRGAMMAI model (). This complete chloroplast genome can be further used for phylogenetic studies of Allium.
Figure 1. Phylogenetic analysis of A. macrostemon with 12 related species. Numbers in the nodes are the bootstrap values.
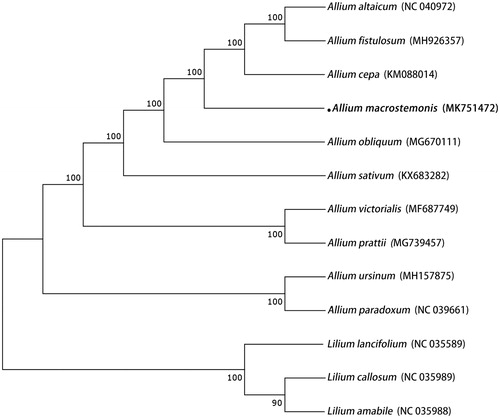
Here, we report complete chloroplast genome of A. macrostemon is 153,158 bp in length, and had a pair of inverted repeats (IR, 26,429 bp, GC – 42.7%) separated by a large (LSC, 82,700 bp, GC–34.6%) and small single-copy (SSC, 17,600 bp, GC – 29.1%) regions. A total of 131 genes were annotated successfully, including 85 protein-coding genes, 8 rRNAs, and 38 tRNAs.
Acknowledgements
The authors would like to thank Xin Yang for helping to complete data analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kuang DY, Wu H, Wang YL, Gao LM, Zhang SZ, Lu L. 2011. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): implication for DNA barcoding and population genetics. Genome. 54:663–673.
- Li F, Xu Q, Zheng T, Huang F, Han L. 2014. Metabonomic analysis of Allium macrostemon bunge as a treatment for acute myocardial ischemia in rats. J Pharm Biomed Anal. 88:225–234.
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci USA. 107:4623–4628.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
