Abstract
The first complete chloroplast (cp) genome of the Chinese endemic and endangered species, Magnolia omeiensis, were reported in this study. The M. omeiensis cp genome was 160,091 bp in length, with two inverted repeat (IR) regions of 26,584 bp, one large single-copy (LSC) region of 88,160 bp, and one small single-copy (SSC) region of 18,763 bp. The cp genome of this species contained 113 unique genes, including 79 protein-coding genes, four ribosomal RNA, and 30 transfer RNA genes. The overall GC content was 39.3%. Phylogenetic analysis of 24 representative cp genomes within the genus Magnolia suggested M. omeiensis is closely related to M. yunnanensis.
Magnolia omeiensis (W. C. Cheng) Dandy is an evergreen in the family Magnoliaceae. It is endemic in China and only distributed in Emei Mountain, Sichuan Province, and usually grows from 1,150 to 1,350 m above sea level (Chen and Ma Citation2007). Magnolia omeiensis has large, colourful, and fragrant blooms, and commonly considered as the excellent ornamental tree species (Chen et al. Citation2015). Meanwhile, its flowers and leaves contain the aromatic essential oil, which confers this species an important potential for applications in cosmetic and perfumery industry (Chen et al. Citation2015). At present, M. omeiensis is listed as the First-Grade State Protection plant of China due to few individuals in the native range. To promote the conservation of this species, we sequenced and analysed the complete chloroplast (cp) genome of M. omeiensis using high-throughput sequencing technology.
The fresh leaf of M. omeiensis was collected from Kunming Botanical Garden, Yunnan province, Southwest of China. The voucher specimen (lpssy0025) were deposited in the herbarium of the Liupanshui Normal University (LPSNU). The total DNA was extracted with the cetyl trimethylammonium bromide (CTAB) method (Doyle and Doyle Citation1987) and used for the library construction and sequencing on the Illumina HiSeq 2500 Platform (Illumina, San Diego, CA). Approximately 2 GB raw data were generated and used to de novo assemble the complete cp genome using the GetOrganelle pipeline (https://github.com/Kinggerm/GetOrganelle). All genes encoding proteins, transfer RNAs (tRNAs), and ribosomal RNAs (rRNAs) were automatically annotated using Dual Organellar Genome Annotator (DOGMA, Wyman et al. Citation2004) with manual adjustments. Moreover, the annotated tRNA genes were further confirmed using the online tRNAscan-SE Search Service platform (Lowe and Chan Citation2016). The complete cp genome sequence of M. omeiensis was deposited in GenBank under the accession number MK728935.
The cp genome of M. omeiensis is a typical quadripartite structure and 160,091 bp in size, including a pair of inverted repeats (IRs) of 26,584 bp, a large single-copy (LSC) region of 88,160 bp and a small single-copy (SSC) region of 18,763 bp. The GC content of the cp genome is 39.3%, similar to the other reported cp genomes from the family of Magnoliaceae (Hinsinger and Strijk Citation2017; Gao et al. Citation2018; Song et al. Citation2018; He et al. Citation2019). The GC content of the LSC, SSC, and IR are 38.0, 34.3, and 43.2%, respectively. The cp genome of M. omeiensis encodes 113 identified genes, including 79 protein-coding genes, 30 transfer RNAs (tRNAs), and 4 ribosomal RNAs (rRNAs), of which 17 genes are duplicated in the IR. Fifteen distinct genes (trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC, atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, and rps16) contain a single intron, while ycf3 and clpP contain two introns.
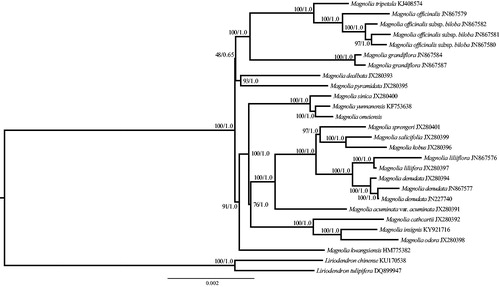
To determine the phylogenetic position of M. omeiensis, a phylogenomic analysis was carried out with the maximum likelihood (ML) and Bayesian inference (BI) methods (Ronquist et al. Citation2012; Stamatakis Citation2014). Two species (Liriodendron chinense and L. tulipifera) were used as the outgroups. The cp genomes of M. omeiensis and previously released cp genomes of the genus Magnolia in NCBI were used for phylogenetic analysis, and the GenBank accession numbers of these species are provided in . All the complete cp genome sequences were aligned using MAFFT version 7.0 (Katoh and Standley Citation2013). The ML and BI analyses generated the same tree topology (). The cp genome data are consistent with morphology, supporting that M. omeiensis was closely related to M. yunnanensis. The M. omeiensis cp genome reported in this study may provide useful resources for the development of ornamental and ecological value as well as robust phylogenetic study at a deep level of Magnolia in the future.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen J, Fan J, Lu C, Zhao B. 2015. Biological characteristics and conservation measures of endangered species Parakmeria omeiensis. Agric Sci Tech Sichuan. 4:22–25.
- Chen Y, Ma M. 2007. Advances in researches on germplasm resources of Parakmeria plants and their propagation technology. J Sichuan Sci Tech. 28:28–33.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Gao J, Song Y, Zheng B. 2018. Complete chloroplast genome sequence of an endangered tree species, Magnolia sieboldii (Magnoliaceae). Mitochondrial DNA Part B. 3:1261–1262.
- He S-L, Wu H-Z, Yang Y. 2019. Characterization of the complete chloroplast genome of Magnolia odoratissima (Magnoliaceae), an endangered and endemic species in China. Mitochondrial DNA Part B. 4:387–389.
- Hinsinger DD, Strijk JS. 2017. The chloroplast genome sequence of Michelia alba (Magnoliaceae), an ornamental tree species. Mitochondrial DNA Part B. 2:9–10.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Song E, Park S, Park J, Kim S. 2018. The chloroplast genome sequence of Magnolia kobus DC. (Magnoliaceae). Mitochondrial DNA Part B. 3:342–343.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.