Abstract
Myripnois dioica (Asteraceae) is a dioecious shrub endemic to North China. In this study, its complete chloroplast genome sequence was assembled and characterized using Illumina paired-end sequencing technology. The whole genome was 153,793 bp in length, containing a pair of inverted repeat (IR) regions of 25,049 bp, a small single-copy (SSC) region of 18,683 bp, and a large single-copy (LSC) region of 85,012 bp. The genome encoded 134 genes including 89 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The overall GC content of the genome was 37.6%. Phylogenetic analysis based on maximum likelihood method indicated Myripnois dioica was close to the taxa in the genus Ligularia. Our results would benefit the conservation genetics and phylogenetic studies of this endemic species.
Myripnois dioica (Asteraceae), the only species in the genus Myripnois, is a dioecious woody shrub endemic to north China (Shi et al. Citation2011). Female and male individuals of M. dioica have purple and white floret corollas, respectively. It is of great horticultural values for its distinctive flower features in early spring and is also used in folk medicine to treat fever (Liu et al. Citation2016). Recently, its population has decreased due to deforestation and overexploitation of natural resources. Myripnois dioica is listed as an endangered species at the provincial level (Wang et al. Citation2015). Conservation and restoration measures should be taken to protect this endemic species. An improved understanding of genetic background is fundamental to formulate effective conservation strategies. However, the present studies on M. dioica are mainly involved in the phylogenetic relationships among taxa in Asteraceae (Funk et al. Citation2014). Little is known about its population genetics information due to the lack of genetic markers. Here, we report the complete chloroplast (cp) genome of M. dioica generated by the Illumina paired-end sequencing technology.
Myripnois dioica was sampled from Baihua Mountain (Beijing, 115.72°E, 39.83°N), China and the voucher (ZP2015BJ27) was deposited at the Evolutionary Botany Laboratory (EBL), Northwest University. Genomic DNA was extracted from leaf tissue with DNAquick Plant System (Tiangen Biotech (Beijing) CO. LTD) and sequenced using the Illumina HiSeq 2500 platform in Biomarker Technologies CO, China. The raw reads were trimmed and assembled following the previously reported method (Peng et al. Citation2019) with Artemisia frigida (NC_020607.1) as the reference. Then, the cp genome was annotated using Geneious R v9.0.5 (Kearse et al. Citation2012).
The complete cp genome of M. dioica (GenBank accession No. MK784068) was 153,793 bp in length and contained a large single-copy (LSC) region of 85,012 bp, a small single-copy (SSC) region of 18,683 bp, and a pair of inverted repeat (IR) regions of 25,049 bp. The plastome contained 134 genes including 87 protein-coding genes, 37 tRNA genes, 8 rRNA genes, and 2 pseudogenes (ycf1, rps19). Among these genes, 10 (rps16, rpoC1, atpF, ndhA, ndhB, petB, petD, rpl16, rpl2 and rps12) harbored one intron and two genes (clpP and ycf3) had two introns. Most genes had a single copy while six protein-coding genes (ndhB, rpl2, rpl23, rps7, ycf2, and ycf15), seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and four rRNA genes (rrn4.5, rrn5, rrn16, and rrn23) were duplicated in the IR regions. The overall GC content was 37.6% and the corresponding values in the LSC, SSC, and IR region was 35.8%, 31.1%, and 43.1%, respectively.
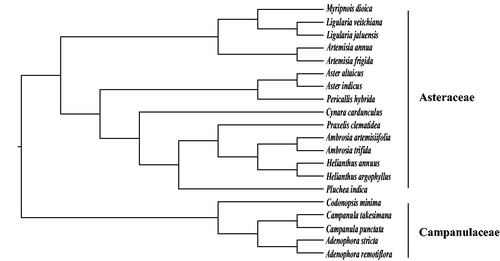
Fourteen published plastomes of Asteraceae were selected to construct the maximum likelihood tree using the reported method (Peng et al. Citation2019) with five Campanulaceae species as outgroup. The phylogenetic tree showed that M. dioica was closely related to Ligularia species (). Our data are helpful for the future systematics and conservation genetics works of M. dioica.
Figure 1. The phylogenetic tree constructed by maximum likelihood method with 20 complete chloroplast genome sequences. Bootstrap support values of all branches are 100%. Accession numbers: Myripnois dioica MK784068, Ligularia veitchiana NC_039385, Ligularia jaluensis NC_039383, Artemisia annua MF623173, Artemisia frigida NC_020607, Aster altaicus NC_034996, Aster indicus NC_040126, Pericallis hybrida NC_031898, Cynara cardunculus KM035764, Praxelis clematidea NC_023833, Ambrosia artemisiifolia MG019037, Ambrosia trifida NC_036810, Helianthus annuus KU306406, Helianthus argophyllus NC_030275, Pluchea indica NC_038194, Codonopsis minima NC_036311, Campanula takesimana NC_026203, Campanula punctata NC_033337, Adenophora stricta NC_036223, Adenophora remotiflora NC-026999.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Funk VA, Sancho G, Roque N, Kelloff CL, Ventosa-Rodriguez I, Diazgranados M, Bonifacino JM, Chan R. 2014. A phylogeny of the Gochnatieae: understanding a critically placed tribe in the Compositae. Taxon. 63:859–882.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxt S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Liu B, Xie Y, Li X, Sun Y, Xu F, Ji T. 2016. [Chemical constituents of Myripnois dioica]. Zhongguo Zhong Yao Za Zhi. 41:3260–3264.
- Peng FF, Chen L, Yang Q, Tian B, Liu ZL. 2019. The complete chloroplast genome of Dipelta yunnanensis (Caprifoliaceae), a vulnerable plant in China. Mitochondrial DNA Part B. 4:515–516.
- Shi Z, Chen YL, Chen YS, Lin YR, Liu SW, Ge XJ, Gao TG, Zhu SX, Liu Y, Yang QE, et al. 2011. Flora of China. Vol. 20–21 (Asteraceae). Beijing: Science Press; St. Louis: Missouri Botanical Garden Press.
- Wang H, Liu Y, Yuan Y, Li X, Du FG. 2015. Rapid propagation in vitro of Myripnois dioica. Plant Physiol J. 51:2275–2279.
