Abstract
We describe the mitogenome sequence of Pseudeuphausia sinica collected in the adjacent waters of the Yangtze River Estuary. The assembled mitogenome is 16,192 bp in length and consists 13 protein-coding genes, 22 transfer-RNA genes, 2 ribosomal-RNA genes, and 1 non-coding region. The most common start codon for 13 PCGs is ATG and the most common termination codon is TAA. The overall G + C content was only 28.26% in the heavy strand. The result of phylogenetic analysis showed that the relationship of P. sinica was close to the species in the same order.
Pseudeuphausia sinica is a common euphausiid species in coastal waters of East China Sea and western Yellow Sea, which was first described by Wang and Chen (Citation1963). The species belongs to the Family Euphausiidae and the Order Euphausiacea. Due to the appropriate bait source of commercial fishes (Yan et al. Citation2006), and being one of the dominant zooplankton species of Lvsi fishing grounds (Yu et al. Citation2013), P. sinica could be important to the local fishery resource supplement. Previous studies mainly focused on the individual development (Wang Citation1965; Li et al. Citation1994), distribution (Chen et al. Citation2008), and the effects of environmental conditions on its population characteristics (Tao et al. Citation2013). Less research is relative to the sequence of the mitochondrial DNA genes (Lin et al. Citation2004).
In the present study, we collected samples of P. sinica in northeast of the Yangtze River Estuary (122°30.8′E, 31°30.2′N; 122°59.6′E, 31°30.6′N) in October 2018 with WP2 net (mesh size: 200 μm). The sample was immediately frozen in –80 °C on board until it was picked out under stereo microscope (Leica S8APO) for mitogenome analysis. The mtDNA was sequenced by Illumina Hiseq 4000. Some samples of P. sinica collected at the same two stations were preserved in 5% formalin solutions and stored in the plankton laboratory of the First Institute of Oceanography, Ministry of Natural Resources. At present, we describe the complete mitochondrial genome of P. sinica, which will help to understand the phylogenetic status of genus Pseudeuphausia among the Class Malacostraca and Phylum Arthropoda.
The length of the complete mitochondrial genome of P. sinica is 16,192 bp with the GenBank accession No. MK579299. The genome contains 37 genes, including 13 protein-coding genes (PCGs), 22 tranfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and 1 control region (D-loop) which is 1518 bp in length. For the 13 PCGs, the most common start codon is ATG (ATP6, COX3, ND4, ND4L and Cytb), then is ATA (COX2, ATP8, ND1 and ND6); the most common termination codon is TAA (COX1, ATP6, ND1, ND2, ND4L and ND6), then is the incomplete termination codon T–– (COX2, COX3, ATP8 and ND4).
The mitochondrial base composition is A 38.37%, T 33.36%, G 11.67%, and C 16.59% in the heavy strand, with an obvious (A + T) % > (G + C) %. Similar situation occurred in the non-coding region (D-loop) in which the (A + T) % was more than 80%.
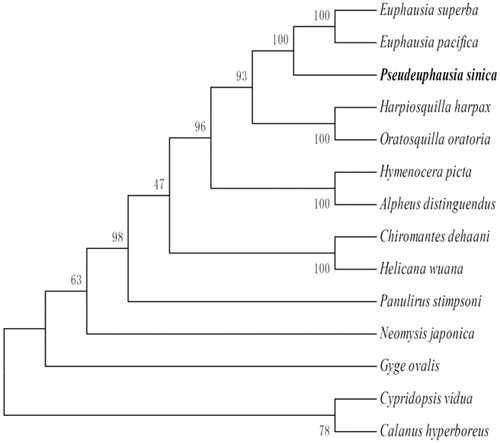
The phylogenetic relationship were estimated using the Maximum Likelihood method in RAxML 8.1.5. It is showed that the phylogenetic relationship of P. sinica is very close to the two species in the Family Euphausiidae: Euphausia pacifica and E. superba. Meanwhile, the phylogenetic relationship of P. sinica is far away from Cypridopsis vidua and Calanus hyperboreus, which are not the species of Class Malacostraca ().
Figure 1. Phylogenetic relationship of 13 species in Phylum Arthropoda based on the concatenated data set of 13 protein-coding genes. Genbank accession Numbers: E. superba (NC016184.1), E. pacifica (EU587005.1), Hymenocera picta (NC039631.1), Oratosquilla oratoria (GQ292769.1), Hymenocera picta (NC039631.1), Alpheus distinguendus (GQ892049.1), Chiromantes dehaani (MH593563.1), Helicana wuana (MH593562.1), Panulirus stimpsoni (GQ292768.1), Neomysis japonica (NC_027510.1), Gyge ovalis (NC037467.1), C. vidua (NC028407), and C. hyperboreus (NC019627.1).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen JJ, Xu ZL, Zhu DD. 2008. Seasonal abundance and distribution of pelagic Euphausiids in the Changjiang Estuary, China. Acta Ecol Sinica. 28:5279–5285.
- Li SJ, Chen F, Wang GZ. 1994. The experimental studies on the growth rate of Pseudeuphausia sinica. J Xiamen Univ (Nat Sci). 33:129–134.
- Lin YS, Cao WQ, Fang LP, Liu QQ, Li SJ. 2004. Analysis of mitochondrial DNA COI gene fragment of Pseudeuphausia sinica. J Xiamen Univ (Nat Sci). 43:842–846.
- Tao ZC, Li CL, Sun S. 2013. The population characteristics and distribution of Pseudeuphausia sinica and the relationship with environmental factors in the Yellow Sea. J Fish China. 37:1782–1794.
- Wang R. 1965. On the larval stages of Pseudeuphausia sinica Wang and Chen (Euphausiacea). Oceanol Limnol Sin. 7:35–58.
- Wang R, Chen KZ. 1963. Description of a new species of the genus Pseudeuphausia (Crustacea)–Pseudeuphausia sinica, sp. nov. Oceanol Limnol Sin. 5:353–359.
- Yan LP, Li JS, Shen DG, Yu LF, Ling LY. 2006. Variations in diet composition and feeding intensity of small yellow croaker Larimichthys polyactis Bleeker in the southern Yellow Sea and northern East China Sea. Mar Fish. 28:117–123.
- Yu WW, Liu PT, Gao YS, Gao JX, Wu FQ, Tang JH, Wu L, Shi JJ. 2013. Distribution characteristics of zooplankton in National Aquatic Germplasm Conversation Area in Lvsi fishing ground of East China Sea in spring and summer. Chin J Ecol. 32:2744–2749.
