Abstract
The mitochondrial genome of Papilio machaon annae is described in this study. The molecule is 15,339 bp in length and contains 13 protein-coding genes (PCGs), two rRNA genes, 22 tRNA genes, and an AT-rich region. All PCGs use the typical start and stop codons, except COI, COII use single T–– and ND3 uses ATG as their stop codons. The rrnL and rrnS genes are 1365 bp and 780 bp in length, respectively. The AT-rich region contains several structures characteristic of the lepidopterans. Phylogenetic analysis shows that P. machaon annae is the closest realtive of Papilio hospiton, rather than the Papilio machaon.
The taxonomic status of Papilio machaon annae Gistel, which is distributed in high latitude areas of northwest China, is still standing as a controversial issue and need to be elucidated. Based on its morphological characteristics, living environment, and host plants, Lee suggested that it should be considered as an effective species while Chou proposed that it is a subspecies of Papilio machaon for the shorter hindwing tail (Lee Citation1980; Chou Citation1994).
Insect mitogenomes have been used as an informative molecular marker for phylogenetic and population genetic studies at various hierarchical levels, due to their unique features (Li et al. Citation2001; Salvato et al. Citation2008). In this study, we newly determined the complete mitochondrial genome of P. machaon annae Gistel. Adult individuals of P. machaon annae Gistel were collected at Qilianshan Mountains, Qinhai province, China (coordinates: N38°5′, E100°39′) on July 2016. After morphological identification, fresh samples were preserved in 100% ethanol and kept in the Laboratory of Molecular Phylogenetics and Evolution (Anhui Normal University, China) at −20 °C until use. Total genomic DNA was extracted from the thorax muscle of a single individual using the Sangon Animal genome DNA Extraction Kit (Shanghai, China). Whole mitochondrial genome was amplified with 14 pairs of primers (Shi et al. Citation2015). The resultant reads were assembled and annotated using the BioEdit 7.0 (Hall Citation1999) and MEGA6 softwares (Tamura et al. Citation2013).
The genome is a circular molecule of 15,339 bp in size (GenBank accession No. MK602655) with a AT bias of 80.8% and contains 13 protein-coding genes (PCGs), 22 tRNA genes, and two rRNA genes and a non-coding AT-rich region (D-loop) as other typical lepidopterans (Feng et al. Citation2010; Hou et al. Citation2016). Thirteen PCGs is 11,195 bp in total, encoding 3720 amino acids. All predicted tRNAs harbor the cloverleaf secondary structures except for the tRNASer (AGN) lacking the DHU-arm (Schattner et al. Citation2005). All protein-coding genes start with a typical ATN codon, except COI which is started with CGA; 10 PCGs stop with usual codon TAA, while COI and COII stop with single T– –, ND3 ends with ATG. The rrnS and rrnL genes are 780 bp and 1365 bp in length, respectively. The AT-rich region is 505 bp in size harboring some structures characteristic of lepidopterans, such as a poly-T stretch upstream of the motif ATAGA, the microsatallite-like elements (TA)6 proceded by the ATTTA motif.
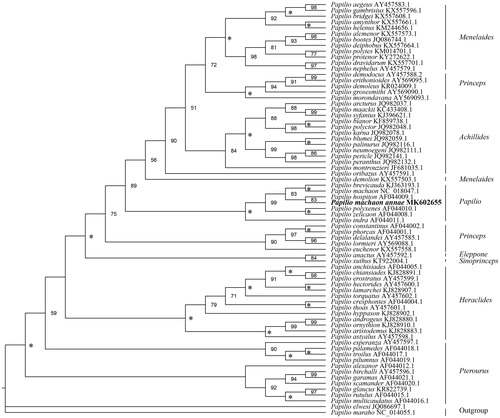
Using Chilasa elwesi and Chilasa maraho as the outgroups, we performed the maximum-likelihood (ML) phylogenetic analysis to clarify the phylogeny of 69 Papilio species, including the P. machaon annae Gistel, based on nucleotide sequence data of COI genes with IQ-TREE (version 1.6.1) ( for details). The resultant ML tree shows that the genus Papilio includes eight subgenera (). The P. machaon annae Gistel is the closest relative of P. hospiton which is distributed in the Mediterranean areas. Thus, P. machaon annae Gistel should be taken as an effective species, rather than as a subspecies of P. machaon.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this paper.
Additional information
Funding
References
- Chou I. 1994. Monographia Rhopalocerorum Sinensium, monograph of Chinese butterflies, Vol. 1. Zhenzhou (China): Henan Scientific & Technological Publishing House.
- Feng X, Liu DF, Wang NX, Zhu CD, Jiang GF. 2010. The mitochondrial genome of the butterfly Papilio xuthus (Lepidoptera: Papilionidae) and related phylogenetic analyses. Mol Biol Rep. 37:3877–3888.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Paper Presented at the Nucleic Acids Symposium Series, 41, 95–98.
- Hou LX, Ying S, Yang XW, Yu Z, Li HM, Qin XM. 2016. The complete mitochondrial genome of Papilio bianor (Lepidoptera: Papilionidae), and its phylogenetic position within Papilionidae. Mitochondrial DNA. 27:102.
- Li M, Badger JH, Chen X, Kwong S, Kearney P, Zhang H. 2001. An information-based sequence distance and its application to whole mitochondrial genome phylogeny. Bioinformatics. 17:149–154.
- Lee CL. 1980. A revision of the Chinese species of Papilio machaon L. and their geographical distribution. Acta Entomol Sin. 23:427–431.
- Salvato P, Simonato M, Battisti A, Negrisolo E. 2008. The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae). BMC Genomics. 9:331.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:686–689.
- Shi QH, Sun XY, Wang YL, Hao JS, Yang Q. 2015. Morphological characters are compatible with mitogenomic data in resolving the phylogeny of nymphalid butterflies (Lepidoptera: Papilionoidea: Nymphalidae). PLoS One. 10:e0124349.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis, version 6.0. Mol Biol Evol. 30:2725–2729.