Abstract
Pomegranate (Punica granatum), as one of the most important fruit trees, is widely cultivated in the tropical and subtropical of the world. In our present study, we assembled a complete chloroplast genome of an important pomegranate cultivar ‘Bhagwa’ using whole genome sequencing data previously reported. The complete cp genome is 158,641 bp in length. A total of 112 unique genes were annotated, including 78 protein-coding genes, 30 tRNA genes, and four rRNA genes. Phylogenetic analysis based on complete cp genomes of Myrtales supported that P. granatum belongs to the Lythraceae family.
Pomegranate (Punica granatum) is one of the oldest known edible fruit trees with a wide geographical global distribution (Teixeira da Silva et al. Citation2013). It is high value for its nutritional, ornamental, and medicinal properties (Zhang et al. Citation2019). As important resources for both basic research and crop breeding, the nuclear genome of P. granatum has been determined (Yuan et al. Citation2018). Compared to the nuclear genome, chloroplast (cp) genome is a low-cost and efficient way to get valuable information for understanding the evolutionary history of plants due to its relatively small size, predominantly uniparental inheritance, and near absence of recombination (Gitzendanner et al. Citation2018; Wang et al. Citation2018). In some cases, high-copy numbers of the cp genome may be found in the whole genome sequencing data of plants, which makes it possible to obtain complete cp genome using whole genome sequencing data (Twyford and Ness Citation2017). Here, we assembled the complete cp genome sequence of pomegranate cultivar ‘Bhagwa’ based on whole genome sequencing data previously reported. Leaf tissues were collected from the adult tree growing in Solapur, India. Total genomic DNA was extracted from leaf tissue from the Bhagwa cultivar. The whole genome sequencing data have been deposited in NCBI with BioSample numbers SAMN08803844.
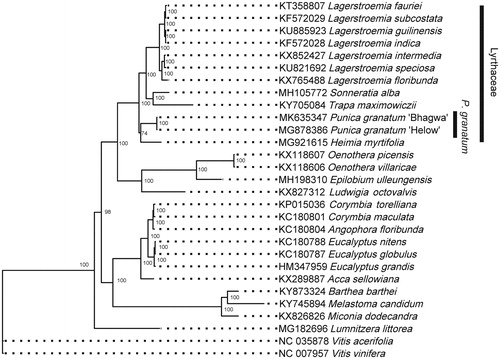
Figure 1. Phylogenetic position of P. granatum within order Myrtales. Phylogenetic tree was constructed using ML method. Numbers in the nodes were the support values.
The raw data (accession number: SRR6917658) was downloaded from the Sequence Read Archive database using ‘fastq-dump’ command. After filtered and trimmed by fastp (Chen et al. Citation2018), high-quality paired-end reads were assembled into a complete cp genome using GetOrganelle (Jin et al. Citation2018). Genome annotation was performed with GeSeq (Tillich et al. Citation2017). The annotation result was inspected using Geneious and adjusted manually as needed. The annotated chloroplast genome has been submitted to GenBank under the accession number MK635347.
The cp genome was 158,641 bp in size with a typical quadripartite, consisting of a pair of inverted repeats (IRs, 25,467 bp), a large single copy (LSC, 89,026 bp), and a small single copy (SSC, 18,682 bp). There were 112 unique genes totally, including 78 protein-coding genes, 30 tRNA genes, and four rRNA genes. Six protein-coding genes (rps7, rps12, rpl2, rpl23, ndhB, ycf2), seven tRNA genes (trnI-CAU, trnN-GUU, trnR-ACG, trnA-UGC, trnI-GAU, trnV-GAC, trnL-CAA), and all rRNA genes (4.5S, 5S, 16S, 23S) are located at the IR regions. The GC content of complete cp genome was 36.92%. The GC content of IRs (42.78%) was higher than that of LSC (34.89%) and SSC (30.64%).
Based on complete cp genome sequences of order Myrtales, the phylogenetic position of P. granatum was inferred using the Maximum likelihood (ML) method. The alignment was achieved by MAFFT (Katoh and Standley Citation2013), and the phylogenetic tree was constructed using IQ-tree (Nguyen et al. Citation2015). The best-fitted model was selected by ModelFinder (Kalyaanamoorthy et al. Citation2017). The result () indicated that P. granatum forms a single clade with other Lythraceae species with high support value. Our result supported that P. granatum belongs to the Lythraceae family.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34:274100.
- Gitzendanner M, Soltis P, Wong G, Ruhfel B, Soltis D. 2018. Plastid phylogenomic analysis of green plants: a billion years of evolutionary history. Am J Bot. 105:291–301.
- Jin J, Yu W, Yang J, Song Y, Yi T, Li D. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv.256479.
- Kalyaanamoorthy S, Minh B, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Nguyen L, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Teixeira da Silva JA, Rana TS, Narzary D, Verma N, Meshram DT, Ranade SA. 2013. Pomegranate biology and biotechnology: a review. Sci Hortic. 160:85–107.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Twyford AD, Ness RW. 2017. Strategies for complete plastid genome sequencing. Mol Ecol Resour. 17:858–868.
- Wang X, Cheng F, Rohlsen D, Bi C, Wang C, Xu Y, Wei S, Ye Q, Yin T, Ye N. 2018. Organellar genome assembly methods and comparative analysis of horticultural plants. Horti Res. 5:3.
- Yuan Z, Fang Y, Zhang T, Fei Z, Han F, Liu C, Liu M, Xiao W, Zhang W, Wu S, et al. 2018. The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol J. 16:1363–1374.
- Zhang T, Liu C, Huang X, Zhang H, Yuan Z. 2019. Land-plant phylogenomic and pomegranate transcriptomic analyses reveal an evolutionary scenario of CYP75 genes subsequent to whole genome duplications. J Plant Biol. 62:48–60.
