Abstract
The complete mitochondrial genome of Otaria byronia was sequenced for the first time. The mitogenome is 16,640 bp long and encodes 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, one origin of light strand replication (OL), and a control region (CR). The overall base composition is 33.1%, 27.0%, 14.2%, and 25.7% for A, C, G, and T, respectively. The CR with 1186 bp is in the position between tRNA-Phe and tRNA-Pro genes. Phylogenetic analyses show the classification status of the pinniped species and reveal that O. byronia is grouped to the Family Otariidae.
The South American sea lion Otaria byronia is mainly distributed along the coasts and offshore islands of South America (Folkens Citation2002). Being the only member of the genus Otaria, this species is now listed as Least Concern status by the International Union for Conservation of Nature (IUCN). As there still exist concerning on the conservation genetics, phylogenetics and evolution of O. byronia, it is necessary to generate its complete mitochondrial genome to provide essential genetic resources for this species.
In the present study, the complete mitochondrial genome of O. byronia is determined and deposited in NCBI GenBank under the Accession Number MK654904. The blood sample was collected from an adult male O. byronia bred in Dalian Sun Asia Tourism Holding Co., Ltd. (GPS location: 38°52′41″N, 121°34′1″E) and stored in a disodium EDTA tube (No. DL-190306) at −80 °C by the Conservation Biology Laboratory of Liaoning Ocean and Fisheries Science Research Institute. Total genomic DNA was extracted using the E.Z.N.A. Tissue DNA Kit (D3396-02) following the standard operation protocol. The whole mitogenome was amplified with 20 pairs of primers (Shanghai Sangon Biotechnology Co., Ltd.). The sequence fragments were assembled using DNAMAN software (https://www.lynnon.com/dnaman.html) and annotated using the program MOTIS search serve (Bernt et al. Citation2013). The transfer RNA (tRNA) genes were recognized using the program tRNAscan-SE search serve (Lowe and Eddy Citation1997). MEGA 7.0 (https://www.megasoftware.net/) was used to perform Maximum-Likelihood (ML) analyses.
The complete mitochondrial genome of O. byronia is 16,640 bp in length, and contains 13 protein-coding genes (PCGs), 22 tRNA genes, two ribosomal RNA (rRNA) genes, an origin of the light-strand replication (OL), and a control region (CR), which is almost identical to those of other marine mammals (Arnason et al. Citation1991; Arnason and Johnsson Citation1992). The total nucleotide composition is 33.1% A, 25.7% T, 27.0% C, and 14.2% G. Most genes are encoded on the heavy strand while ND6 and eight tRNA genes are located on the light strand. The total PCGs of O. byronia (11,427 bp) encode 3798 amino acids. All 22 tRNAs have a potential to fold into the typical cloverleaf structures except for tRNA-Ser (GCT), which loses the stem of dihydrouridine (DHU) arm. It has been reported that the absence of the DHU arm is a characteristic of the gene tRNA-Ser (GCT) among the mammals (Ge et al. Citation2018). 12S rRNA and 16S rRNA, located between tRNA-Phe and tRNA-Leu and separated by the tRNA-Val, are 961 and 1578 bp, respectively. The OL between tRNA-Asn and tRNA-Cys is 31 bp in length, and the CR (1186 bp) is located between tRNA-Pro and tRNA-Phe.
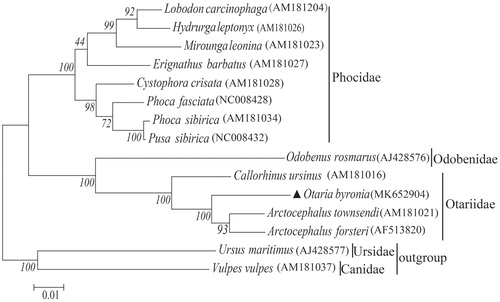
Phylogenetic analyses were carried out based on 12 pinniped species closely related to O. byronia by the maximum likelihood (ML) methods (). The phylogenetic tree was rooted with Ursus maritimus and Vulpes vulpes. It provides strong bootstrap support for the relationship between Otariidae, Odobenidae, and Phocidae and reveals that O. byronia is grouped to the Family Otariidae, which is consistent with paleontological and morphological findings of Otariidae evolution (Repenning et al. Citation1979; Brunner Citation2004).
Acknowledgments
The authors would like to thank the vet of the Dalian Sun Asia Tourism Holding Co., Ltd. for his assistance in sample collection.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Arnason U, Gullberg A, Widegren B. 1991. The complete nucleotide sequence of the mitochondrial DNA of the fin whale, Balaenoptera physalus. J Mol Evol. 33:556–568.
- Arnason U, Johnsson E. 1992. The complete mitochondrial DNA sequence of the harbor seal, Phoca vitulina. J Mol Evol. 34:493–505.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. Mitos: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Brunner S. 2004. Fur seals and sea lions (Otariidae): identification of species and taxonomic review. Syst Biodivers. 1:339–439.
- Folkens P. 2002. National Audubon society guide to marine mammals of the world. New York: Alfred A. Knopf, Chanticleer Press Edition.
- Ge Y, Zhu L, Chen M, Zhang G, Huang Z, Cheng R. 2018. Complete mitochondrial genome sequence for the endangered Knysna seahorse Hippocampus capensis Boulenger 1900. Conserv Genet Resour. 10:461–465.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Repenning CA, Ray CE, Grigorescu D. (1979). Pinniped biogeography. In: Gray J, Boucot AJ, editors. Historical biogeography, plate tectonics, and the changing environment. Oregon: Oregon State University Press.