Abstract
Aenasius arizonensis is the endoparasitoid of mealybug (Phenacoccus solenopsis) and plays an important role in biological control. We sequenced the A. arizonensis mitochondrial genome using next-generation sequencing. The mitochondrial genome of A. arizonensis is 15,373 bp in length with a typical 37 gene pattern. The base composition is AT-biased (79.61%). Both PCGs and tRNA gene rearrangement events are detected. Six PCG inversion events are detected in the mitochondrial genome of A. arizonensis. The tRNA gene shuffling between nad2 and nad3 was detected for the first time in Chalcidoidea mitochondrial genomes. The phylogenetic analysis supported the monophyly of Chalcidoidea. The sister group of (Trichogrammatidae + Encyrtidae) and Pteromalidae was corroborated by both Bayesian and maximum likelihood methods.
Aenasius arizonensis (Hymenoptera: Encyrtidae) is a kind of important endoparasitoid of mealybug, Phenacoccus solenopsis (Hemiptera: Pseudococidae). Aenasius arizonensis is among the most important groups of natural enemies of scale insects and have been utilized extensively in biological control (Noyes Citation2018). The species of Encyrtidae attack a wide range of insect hosts. However, the species with sequenced complete or nearly complete mitogenomes of this family is rare. Here, we present the mitochondrial genome of A. arizonensis.
Voucher DNA and specimens are deposited in the Institute of Insect Sciences, Zhejiang University. Specimens were stored at 4 °C before DNA extraction. The whole genomic DNA was extracted from a male adult specimen using the DNeasy tissue kit (Qiagen, Hilden, Germany). The mitochondrial genome of A. arizonensis was obtained using next-generation sequencing. The library was constructed with VAHTSTM Universal DNA Library Prep Kit for Illumina and sequenced with Illumina Hiseq X Ten sequencer (150 bp paired-end). DNA library was assembled by Celera Assembler v7.0 (Myers et al. Citation2000) and IDBA_tran (Peng et al. Citation2012). We annotated protein-coding genes (PCGs), rRNAs, and tRNAs using MITOS (Bernt et al. Citation2013), and OrfFinder (Sayers et al. Citation2016).
The mitochondrial genome of A. arizonensis is 15,373 bp in length (GenBank accession number: MK630013) and contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs) and a control region (non-coding AT-rich region). The nucleotide composition of the whole mitochondrial genome is significantly biased with A + T contents of 79.61%. All the tRNA genes have typical cloverleaf structure except for trnS1, which has been reported in the mitochondrial genomes of most insects (Wolstenholme Citation1992).
All PCGs start with typical ATN codons (three with ATA, seven with ATT, and three with ATG). Six genes terminate with TAA, three genes terminate with TAG and four genes possess incomplete stop codon. Similar to other parasitoid wasps, massive gene rearrangement events are detected in the mitochondrial genome of A. arizonensis compared with pancrustacean ancestral gene order. Six PCG inversion events (cox1–cox2–atp8–atp6–cox3–nad3) are detected in the mitochondrial genome of A. arizonensis. This phenomenon is common in Chalcidoidea mitochondrial genomes. However, we found a novel tRNA cluster shuffling pattern between nad2 and nad3 compared with other Chalcidoidea mitochondrial genomes. The tRNA gene trnN are relocating between trnW and trnY.
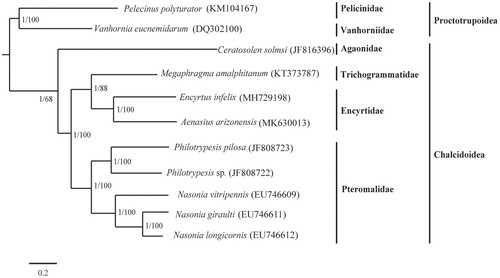
We reconstructed the phylogeny of Chalcidoidea using nine complete or nearly complete mitogenomes of this lineage available in GenBank. We selected Pelecinus polyturator and Vanhornia eucnemidarum as outgroup (). We partitioned the protein-coding genes using PartitionFinder. Two phylogenetic algorithms, Bayesian and Maximum likelihood methods, were used to reconstruct the phylogeny. Two consensus trees showed identical topology. The monophyly of Chalcidoidea was supported. Agaonidae is nested at the basal position of the phylogeny. The sister group of (Trichogrammatidae + Encyrtidae) and Pteromalidae was corroborated by previous analyses (Xiong et al. Citation2019; Zhu et al. Citation2018).
Figure 1. The molecular phylogeny of Chalcidoidea based on partitioned protein-coding sequences. The phylogenetic tree was constructed by Bayesian inference and maximum-likelihood methods under GTRGAMMA model. The number at each node indicate the posterior probability and bootstrap values resulting from the analyses.
Disclosure statement
No potential conflict of interest was reported by the authors. The authors are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogen Evol. 69:313–319.
- Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, Remington KA, et al. 2000. A whole-genome assembly of Drosophila. Science. 287:2196–2204.
- Noyes JS. 2018. Universal Chalcidoidea database. World Wide Web electronic publication. [accessed 2018 June 1]. http://www.nhm.ac.uk/chalcidoids.
- Peng Y, Leung HC, Yiu SM, Chin FY. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28:1420–1428.
- Sayers EW, Agarwala R, Bolton EE, Brister JR, Canese K, Clark K, Connor R, Fiorini N, Funk K, Hefferon T, et al. 2016. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 44:D7.
- Wolstenholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173.
- Xiong M, Zhou Q, Zhang Y. 2019. The complete mitochondrial genome of Encyrtus infelix (Hymenoptera: Encyrtidae). Mitochondrial DNA Part B. 4:114–115.
- Zhu J, Tang P, Zheng B, Wu Q, Wei S, Chen X. 2018. The first two mitochondrial genomes of the family Aphelinidae with novel gene orders and phylogenetic implications. Int J Biol Macromol. 118:386–396.
