Abstract
Helianthemum songaricum is an ecologically important endangered relic plant species growing in the arid and semi-arid regions of northwestern China. It is also a valuable germplasm resource. Consequently, we herein report the complete H. songaricum chloroplast genome, which comprises 152,042 bp, including a large single copy (LSC) region (84,389 bp) and a small single copy (SSC) region (18,926 bp) that are separated by a pair of 24,298-bp inverted repeats (IRs). The LSC, SSC, and IR regions account for 55.50%, 12.45%, and 31.96% of the chloroplast genome, respectively. Moreover, the GC content of this genome is 36.6%. Furthermore, the 110 annotated genes consist of 75 protein-coding genes, 4 rRNAs, and 31 tRNAs. A phylogenetic tree based on 14 chloroplast genomes revealed that H. songaricum is closely related to Clematoclethra scandens and Actinidia arguta.
Helianthemum songaricum (Cistaceae) is a deciduous shrub or subshrub species (Chinese Academy of Science, Chinese Ethnography Editorial Board Citation1990) that grows primarily on lithoid hillsides (1000–1500 m above sea level) in regions with an arid climate (Su et al. Citation2011). Additionally, H. songaricum is an endangered species in the family Cistaceae (Han et al. Citation2017), which comprises eight genera and 200 species that are mostly distributed along the Mediterranean coast but have also been detected in the Americas and Asia (Proctor and Heywood Citation1978; Marrero and Mesa Citation2003). H. songaricum is the only Cistaceae genus identified in China (Gao et al. Citation2006). Specifically, H. songaricum is an ecologically and economically important and endangered relic plant in the arid and semi-arid areas in northwestern China. The fact that H. songaricum is on the verge of extinction is likely because of human activities and many natural factors. Previous research involving horizontal starch gel electrophoresis analyzed the genetic diversity and population differentiation of this species (Duan et al. Citation2000). However, the characteristics of the H. songaricum chloroplast genome (cpDNA) have not been elucidated. In this study, we sequenced, assembled, annotated, and comparatively analyzed the distribution and structural characteristics of the H. songaricum cpDNA. We also completed a phylogenetic analysis of the chloroplast protein-coding gene variations among species and examined the environmental adaptability of H. songaricum. The results of this research may provide a theoretical basis for future investigations of H. songaricum.
Fresh H. songaricum leaves were collected in Qipanjing, Ningxia Hui Autonomous Region, China (107° 07.1560′ E, 39° 21.2386′ N; height: 1,404 m above sea level) in June 2018. The specimens of H. songaricum were deposited at the Herbarium of Yulin University, Shaanxi province, China. Genomic DNA was extracted from the fresh leaves according to a modified CTAB method (Doyle and Doyle Citation1987). The subsequent high-throughput sequencing was completed with the Illumina HiSeq X Ten system, with the Camellia azalea chloroplast genome (NC_035574) used as the reference sequence. The H. songaricum chloroplast genome was annotated with the Geneious program (Biomatters Ltd., Auckland, New Zealand) after its sequence was aligned with the reference chloroplast genome. The OGDraw online tool was used to visualize the H. songaricum chloroplast genome (Lohse et al. Citation2013). We used the MEGA6.0 program (Tamura et al. Citation2013) to construct a phylogenetic tree according to the neighbor-joining method, with a bootstrap value of 1000. The annotated H. songaricum complete chloroplast genome sequence has been deposited into the GenBank database (MK776534).
Our data revealed that the H. songaricum chloroplast genome comprises 152,042 bp, which includes a large single copy region (LSC; 84,389 bp), a small single copy region (SSC; 18,926 bp), and a pair of inverted repeats (IRs; 24,298 bp). Additionally, the genome has a GC content of 36.6%. The LSC, SSC, and IR regions represent 55.50, 12.45, and 31.96% of the H. songaricum chloroplast genome length, respectively. Furthermore, we annotated 110 genes, with 75, 4 and 31 genes encoding proteins, rRNAs, and tRNAs, respectively.
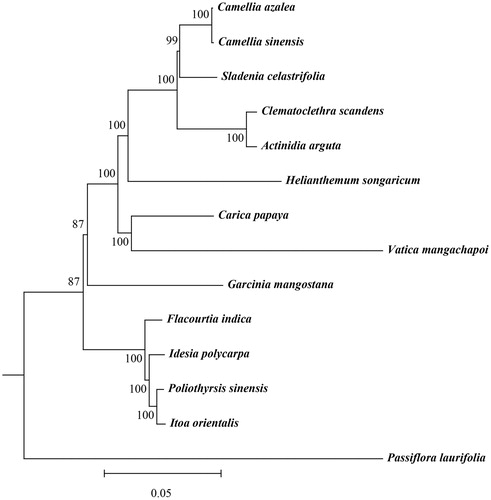
We constructed a phylogenetic tree based on 14 complete chloroplast genomes, with the Camellia sinensis chloroplast genome serving as an outgroup (). The resulting tree revealed that H. songaricum is closely related to Clematoclethra scandens and Actinidia arguta, but is clustered by itself. The tree also clarified the relationships between H. songaricum and other species, thereby providing useful information for future studies of H. songaricum.
Figure 1. Phylogenetic tree constructed based on 14 species of chloroplast genomes. Accession numbers: Flacourtia indica (NC_037410); Poliothyrsis sinensis (NC_037412); Idesia polycarpa (KX229742); Itoa orientalis (MG262342); Carica papaya (EU431223); Vatica mangachapoi (MH716496); Sladenia celastrifolia (NC_035707); Actinidia arguta (MG744576); Clematoclethra scandens (KX345299); Camellia azalea (KY856741); Garcinia mangostana (NC_036341); Passiflora laurifolia (MF807939); Helianthemum songaricum (MK776534); Camellia sinensis (KC143082).
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Chinese Academy of Science, Chinese Ethnography Editorial Board. 1990. Flora reipublicae popularis sinicae, (Vol. 50, Volume 2). Beijing, China: Science Press; p. 178–179. Chinese.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phyt Bul. 19:11–15.
- Duan FZ, Cao R, Ma H, Zhao YZ, Hou X. 2000. Genetic divertisty and differentiation of endangered species—Helianthemum songaricum. China Sci Abstr. 6:217–219.
- Gao TP, Zhang Y, Jin L, Chen T, Xu SJ, An LZ. 2006. Research advances on rare and endangered plants Cistaceae. J Dese Rese. 26: 312–316. Chinese.
- Han BC, Wei LY, Wang XY, Liang XH, Wulanbateer SD. 2017. Study on genetic diversity and phylogeographic structure of endangered Helianthemum soongoricum. J Inner Mongolia Fore Sci and Tech. 43:16–19. Chinese).
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucl Acids Res. 41:W575–W81.
- Marrero A, Mesa R. 2003. The genus Helianthemum Mill. In La Gomera. Cana I Slan Cando. 58:149–162.
- Proctor MCF, Heywood VH. 1978. Flora European (2nd edition). London, UK: Cambridge University Press; p. 286–291.
- Su ZH, Zhang ML, Sanderson SC. 2011. Chloroplast phylogeography of Helianthemum songaricum (Cistaceae) from northwestern China: implications for preservation of genetic diversity. Conservation Gen. 12:1525–1537.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 30:2725–2729.
