Abstract
Elaeagnus conferta Roxb is a kind of fruit known for its oval shape, bright red colour, and sour and sweet taste. In this study, we assembled and characterized the complete chloroplast genome of E. conferta Roxb as a resource for future genetic studies. With a total length of 151,751 bp, the chloroplast genome has a total of 130 genes, consisting of 88 protein-coding genes, 34 tRNA genes, and 8 rRNA genes. Phylogenetic analysis confirmed the position of E. conferta Roxb within the order Rosales.
Elaeagnus conferta Roxb, a kind of peculiar and rare tropical fruit (Ruchang et al. Citation2011) native to Vietnam, Malaya, India, southern China and other tropical Asian regions, belongs to the Elaeagnaceaea family (Yu et al. Citation2016). The fruits of E. conferta Roxb enriched with 8 essential amino acids, lycopene and carotene, have high nutritional value (Zhonghua et al. Citation2018). The root and leaf are traditional plant medicine in southeast Asia, have the effect of astringency, antidiarrheal, antitussive and detoxification (Yumei and Weinan Citation2007). In recent years, the nutritional and medicinal value of E. conferta Roxb has been widely concerned, however, there is little research on the genetics and genomics of E. conferta Roxb. In this study, we report the complete chloroplast genome of E. conferta Roxb, and hope to provide genomics data for the genetic research of wild germplasm resources of E. conferta Roxb.
DNA material was isolated from mature leaves of an E. conferta Roxb plant cultivated in the plant garden of Yunnan Institute of Tropical Crops (YITC), Jinghong, China, by using DNeasy Plant Mini Kit (QIAGEN, Germany) and the specimen of this tree and the isolated DNA were stored in YITC. After DNA extraction, a library with the insertion size of 400 bp was constructed, and high-throughput DNA sequencing (pair-end 150 bp) was performed on an Illumina Hiseq 2500 platform, generating ∼3 Gb of sequence data. Using an E. conferta Roxb partial rbcL gene sequence (Y15139.1) as seed sequence, the chloroplast genome was assembled from the Illumina data using the program NOVOPlasty (Dierckxsens et al. Citation2017). The assembled chloroplast genome sequence was then annotated using DOGMA (Wyman et al. Citation2004) and manually corrected.
The complete chloroplast genome sequence of E. conferta Roxb (GenBank accession MK404307) was 151,751 bp in length, with a large single-copy (LSC) region of 82,533 bp, a small single-copy (SSC) region of 18,024 bp, and two inverted repeat (IR) regions of 25,597 bp each. A total of 130 genes were predicted, consisting of 88 protein-coding genes, 34 tRNA genes, and 8 rRNA genes. The overall GC content was 37%.
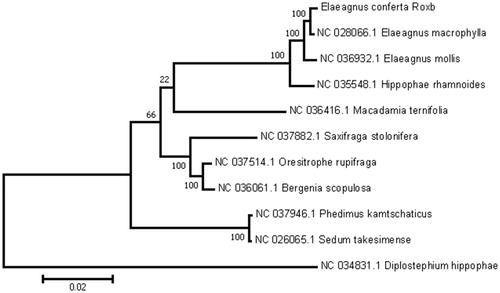
To further investigate its phylogenetic position, a maximum-likelihood tree was constructed based on complete chloroplast genome sequences of 10 other Saliaceae species using MEGA7 (Kumar et al. Citation2016) with 500 bootstrap replicates. Here, we aligned all 10 sequences using MAFFT (Katoh and Standley Citation2013). As shown in , E. conferta Roxb clustered within Rosales with strong bootstrap support.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger Datasets. Mol Biol Evol. 33:1870–1874.
- Ruchang F, Haixing Y, Guanfu L, Dongli N, Sanwei X. 2011. Breeding of Elaeagnus conferta Roxb. Chin J Trop Agric. 31:20–23.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yu G, Minjie D, Jiashui W, Shun S, Xiaoping Z, Xinge L, Weihong M. 2016. The briefing collection and evaluation of Elaeagnus conferta in Yunnan province. Chin J Trop Agric. 36:48–50.
- Yumei L, Weinan H. 2007. Fatty acids and mineral elements of seeds of Elaeagnus conferta Roxb. J Trop Subtrop Bot. 15:253–255.
- Zhonghua L, Qian L, Junbo Z, Ping J, Hongyan T, Jianzhu Z. 2018. Study on cuttage of Elaeagnus conferta Roxb. Shandong Forest Sci Technol. 237:47–49.