Abstract
Discogobio laticeps is a small native economic fish species in southern China. Here, we reported the complete mitochondrial genome of D. laticeps for the first time. It was 16,596 bp in length and composed of 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, a light strand origin of replication (OL), and a control region (D-loop). The overall base composition was 32.3% of A, 15.2% of G, 26.4% of T, and 26.1% of C, with a slight A + T content of 58.7%. Maximum-likelihood phylogenetic tree suggested that D. laticeps had the closest evolutionary relationship with Discogobio longibarbatus.
Discogobio laticeps is a small freshwater fish endemic to southern China, belonging to the subfamily Labeoninae and classified as a new species since 1993. The specialization of the sucker structure on their lip is the adaptation of rapid stream. By now, there are only a few morphological and taxonomic researches about this species (Chu et al. Citation1993, Du et al. Citation2008). The characterization of the complete mitochondrial genome of D. laticeps and its molecular evolution in Labeoninae fish remain unknown. Here, we firstly identified and analyzed the complete mitogenome of D. laticeps and it would be useful to the researches on the origin and phylogeny of this species.
The specimens of D. laticeps were collected from Du'an Yao Autonomous County (23°56′1.55″N 108°06′4.84″E). Muscle samples were fixed in 95% ethanol and stored at −20 °C in Guangxi Colleges and Universities Key Laboratory of Aquatic Healthy Breeding and Nutrition Regulation, Guangxi University. The total genomic DNA was extracted from muscle. Twelve pairs of primers were designed and the sequences were amplified by PCR. Finally, the mitochondrial genome was annotated though MitoAnnotator (Iwasaki et al. Citation2013).
The complete mitochondrial genome of D. laticeps was 16,596 bp in length (GenBank accession no. MH127917), with 58.7% A + T content. The whole base composition was 32.3% of A, 15.2% of G, 26.4% of T, 26.1% of C. Mitochondrial genome organization included 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, a light strand origin of replication (OL), and a control region (D-loop). It showed a high degree of conservation among D. laticeps and other Labeoninae fishes. Except for ND6 and eight tRNA genes on the light strand, most mitochondrial genes were on the heavy strand. Among the 13 PCGs, 12 PCGs shared ATG as the start codon, except for COI which had GTG as the start codon. Besides, there were seven PCGs used the common stop codon of TAA and the remaining six PCGs employed the stop codon of T or TA. The lengths of 12S and 16S rRNA were 950 and 1686 bp, respectively. In addition, the light strand origin of replication (OL) was 36 bp in length and located between tRNAAsn and tRNACys. The control region (D-loop) was 939 bp long and located between tRNAPro and tRNAPhe.
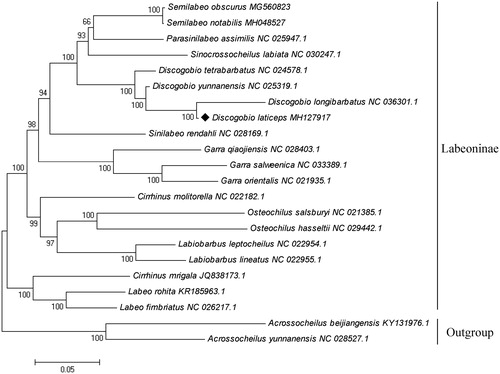
To further investigate the phylogeny of D. laticeps, we constructed the phylogenetic tree based on 13 mitochondrial PCGs of 20 Labeoninae species and chosen two Barbinae species as outgroup by MEGA 7.0 software (Kumar et al. Citation2016). Evolutionary model selection was inferred using PhyML 3.0, in which the substitution model GTR + G + I was considered to be the most suitable model (Guindon et al. Citation2010). The phylogenetic tree showed a sister relationship between D. laticeps and D. longibarbatus (). The results of this study provided a reference for the mitochondrial genome researches of Labeoninae and it would be beneficial to promote the studies of genomics and phylogenetics of this species.
Figure 1. Phylogenetic tree of Discogobio laticeps among 20 Labeoninae species based on 13 concatenated mitochondrial PCGs by maximum-likelihood analysis with GTR + G + I model. Two species were selected to Barbinae as out-groups. The number on branches indicates posterior probabilities in percentage. The number after the species name is the GenBank accession number. The mitogenomic information of Discogobio laticeps is marked with a rhombus.
Disclosure statement
The authors report no conflicts of interest and are solely responsible for this paper.
Additional information
Funding
References
- Chu XL, Cui GH, Zhou W. 1993. A taxonomic review of fishes of the genus Discogobio, with description of two new species (Cypriniformes: Cyprinidae). Acta Zoot Sin. 18:237–246.
- Du LN, Huang YF, Chen XY, Yang JX. 2008. Three new records of fish in yunnan and analysis of the value of faunal presence of fish in the Tuoniang River. Zool Res. 29:69–77.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. Mitofish and Mitoannotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
