Abstract
Platycodon grandiflorus is the single genus plant in family Campanulaceae, which is distributed in areas of China, Japan, Korea, and Russia. In this study, we assembled and annotated the complete chloroplast genome of P. grandiflorus. The chloroplast genome of P. grandiflorus is 171,624 bp long, containing a shorter small single-copy region (SSC) of 7,962 bp, a normal large single-copy region (LSC) of 79,690 bp and a longer pair of inverted repeat regions (IRs) of 41,986 bp. The whole chloroplast genome of P. grandiflorus has 146 genes, including 102 protein-coding genes (PCGs), 36 transfer RNA genes (tRNAs), and 8 ribosome RNA genes (rRNAs). The overall nucleotide composition is: A of 30.9%, T of 30.9%, C of 19.3%, and G of 18.9%, with a total G + C content of the chloroplast genome 38.2%. The phylogenetic and genetic analysis showed a close relationship between Platycodon grandiflorus and Codonopsis lanceolata.
Platycodon grandiflorus is a species of herbaceous flowering perennial plant of the family Campanulaceae, which is the only member of the genus Platycodon plant (Zhang et al. Citation2015). It is native to China, Japan, Korea, and East Asia areas, and has been widely used as traditional medicine and food in these countries (Zhao et al. Citation1994). It is one of the most commonly used traditional Chinese medicine, such as using at anti-tumor, hepatoprotective, immunoregulatory, and anti-oxidant effects (Zhao et al. Citation2017). At present, 100 compounds have been isolated from P. grandiflorus, including triterpenoid saponins, flavonoids, phenolic acids, polyacetylenes, and other compounds (Zhao et al. Citation1994; Zheng et al. Citation2010). Triterpenoid saponins are one of the main active component in P. grandiflorus that can have anti-oxidant, anti-obesity, anti-inflammation and anti-tumor activities (Kim et al. Citation2006). Until now, limited genome information of P. grandiflorus was known. So, we annotated the complete chloroplast genome of P. grandiflorus that analyzed the genetic and phylogenetic relationship and set data of P. grandiflorus in China.
The specimen sample of P. grandiflorus was purchased from Yanji western market (Jinan, Yanji, China, 129.51E; 42.91N). The whole plant tissue and the cpDNA was extracted using the CTAB protocol and stored in Jilin Engineering Normal University Laboratory (Number JLENUL001). The cpDNA was purified and fragmented using the NEB Next UltraTM II DNA Library Prep Kit (NEB, China) that the cp genome was sequenced. The chloroplast genome of P. grandiflorus was assembled and annotated using MitoZ (Meng et al. Citation2019). The tRNA sequences were confirmed using tRNAscan-SE web (Schattner et al. Citation2005). The chloroplast genome map of P. grandiflorus was generated using OrganellarGenomeDRAW (Lohse et al. Citation2013).
The whole complete chloroplast genome of P. grandiflorus (NCBI No.MK6998771) is a circle and the length of 171,624 bp, which is longer than other plants. It has a shorter small single-copy region (SSC) of 7962 bp, a normal large single-copy region (LSC) of 79,690 bp and a longer pair of inverted repeat regions (IRs) of 41,986 bp. The cpDNA of P. grandiflorus comprised 146 genes, including 102 protein-coding genes (PCGs), 36 transfer RNA genes (tRNAs), and 8 ribosomal RNA genes (rRNAs). The IR is longer than other plants and has 32 genes species in one IR, including 21 PCGs species (rpl36, rps8, rpl14, rpl16, rps3, rpl22, rps19, rpl2, rpl23, ycf2, ycf15, ndhB, rps7, rps12, ycf1, rps15, ndhH, ndhA, ndhI, ndhG and ndhE), 7 tRNA species (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG and trnN-GUU) and 4 rRNA species (rrn16S, rrn23S, rrn4.5S and rrn5S). The overall nucleotide composition is: 30.9% of A, 30.9% of T, 19.3% of C and 18.9% of G, with a total G + C content of 38.2%.
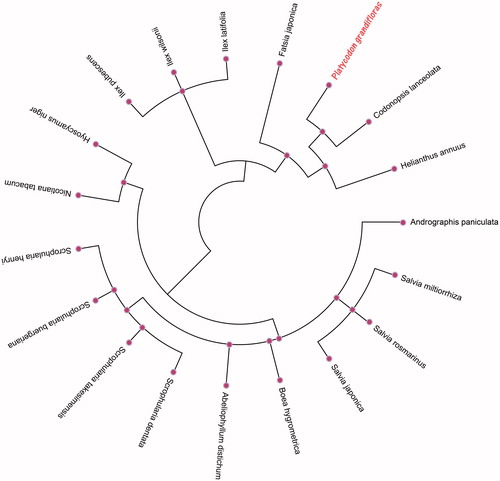
The 18 herbal medicine plant species’ chloroplast genomes were selected to study the phylogenetic relationships with P. grandiflorus. The genetic and phylogenetic tree was reconstructed using the maximum-likelihood (ML) methods analysis. ML analysis was performed using MEGA X (Kumar et al. Citation2018) and inferred using 5000 bootstrap replicates, which was based on the most appropriate model with the GTRCAT substitution model. The ML phylogenetic tree was represented using MEGA X and edited using the iTOL online web (https://itol.embl.de/). As shown in the phylogenetic ML tree (), the chloroplast genome of P. grandiflorus is clustered and closest to the family Campanulaceae of Codonopsis lanceolata (GenBank No. MH018574.1) in the evolutionary relationship. The complete chloroplast genome of P. grandiflorus will provide a useful genome resource for studying the genetic diversity for this species and the family Campanulaceae.
Figure 1. The genetic and phylogenetic tree inferred using maximum-likelihood (ml) analysis from 19 species complete chloroplast genomes of herbal medicine plants. The bootstrap values were based on 5000 replicates. in this study 18 herbal medicine species chloroplast genomes have been deposited in the GenBank and accession numbers are as follows: Abeliophyllum distichum (KT274029.1), Andrographis paniculata (KF150644.2), Boea hygrometrica (JN107811.1), Codonopsis lanceolata (MH018574.1), Fatsia japonica (KR021045.1), Helianthus annuus (DQ383815.1), Hyoscyamus niger (KF248009.1), Ilex latifolia (KX426465.1), Ilex pubescens (KX426467.1), Ilex wilsonii (KX426471.1), Nicotiana tabacum (Z00044.2), Salvia japonica (KY646163.1), Salvia miltiorrhiza (JX312195.1), Salvia rosmarinus (KR232566.1), Scrophularia buergeriana (KP718626.1), Scrophularia dentate (KT428154.1), Scrophularia henryi (MF861203.1), Scrophularia takesimensis (KP718628.1).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Kim JY, Kim DH, Kim HG, Song GY, Chung YC, Roh SH, Jeong HG. 2006. Inhibition of tumor necrosis factor-α-induced expression of adhesion molecules in human endothelial cells by the saponins derived from roots of Platycodon grandiflorum. Toxicol Appl Pharmacol. 210:150–156.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucl Acids Res. 41:W575–W581.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucl Acids Res. doi:10.1093/nar/gkz173.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucl Acids Res. 33:W686–W689.
- Zhang L, Wang Y, Yang D, Zhang C, Zhang N, Li M, Liu Y. 2015. Platycodon grandiflorus-An Ethnopharmacological, phytochemical and pharmacological review. J Ethnopharmacol. 164:147–161.
- Zhao SC, Fu L, Liu ML, Han M. 1994. Determination on the nutrition component of Platycodon grandiflorum etc three kinds of plants. Food Science. 4:47–49.
- Zhao X, Wang Y, Yan P, Cheng G, Wang C, Geng N, Wang X, Liu J. 2017. Effects of polysaccharides from Platycodon grandiflorum on immunity-enhancing activity in vitro. Molecules. 22:1918.
- Zheng J, He J, Ji BP. 2010. Influence of the ethanol fraction from Platycodon grandiflorum on glucose tolerance of streptozotocin-induced diabetic rat. Pharmacol Clin Chin Materia Medica. 26:52–55.
