Abstract
The complete chloroplast genome sequence of a Betulaceae species, Corylus heterophylla var. sutchuenensis Franch, was mapped and determined based on Illumina sequencing data. The complete chloroplast genome is 161,127 bp long and comprises a pair of inverted repeat regions of 26,615 bp each, a large single-copy region of 89,119 bp, and a small single-copy region of 18,778 bp. It harbors 112 genes, including 78 protein-coding genes, 4 ribosomal RNA genes, and 30 transfer RNA genes. Phylogenetic analysis based on chloroplast genomes indicates that C. heterophylla var. sutchuenensis is similar to Corylus chinensis and closely related to Ostrya rehderiana.
Corylus heterophylla var. sutchuenensis, a Chinese endemic hazelnut species, is mainly distributed in the Qilian and Minshan Mountains of Gansu Province, Qinling Mountains of Shanxi Province, Funiu and Dabie Mountains of Henan Province, Daba and Leshan Mountains of Sichuan Province, Heng and Tianmen Mountains of Hunan Province, Shennongjia Mountains of Hubei, and Fanjing Mountains of Guizhou Province, at latitudes of 24.92–36.78°N and longitudes of 98.08–121.3°E (Huo et al. Citation2016). Because of the limited molecular genetics data available, there are different views on the taxonomy of this species, including that C. heterophylla var. sutchuenensis is a separate species (Hu X Citation1955) or just a C. heterophylla variant (Zhang et al. Citation2005). Although several chloroplast (cp) DNA markers have previously been used for the phylogenetic analysis of Corylus (Erdogan and Mehlenbacher Citation2000; Palme and Vendramin Citation2002; Boccacci and Botta Citation2009; Bassil et al. Citation2013; Martins et al. Citation2013), except for Corylus chinensis (GenBank accession number KX814336) (Hu et al. Citation2017), little is known about the cp genome in this genus. In the present study, we report the first complete cp genome sequence of C. heterophylla var. sutchuenensis (GenBank accession number MF996573) based on Illumina paired-end sequencing data.
Fresh leaves were collected from a single C. heterophylla var. sutchuenensis plant growing at the resources nursery of the Beijing Academy of Forestry and Pomology Sciences (Beijing, China). Voucher specimens were deposited at Beijing Academy of Forestry and Pomology Sciences Herbarium. DNA extraction was performed according to a modified CTAB protocol (Li et al. Citation2013). High-throughput sequencing was carried out using the HiSeq4000 PE150 system (Illumina, San Diego, CA, USA). A total of 1,091,337 reads were obtained and used for cp genome assembly with the SPAdes 3.6.1 (Bankevich et al. Citation2012) and SOAPdenovo (Luo et al. Citation2012) software tools. Reference-guided assembly was then performed to reconstruct the cp genome with the BLAST program (Altschul et al. Citation1990) using closely related species as references. After filling the gaps with GapCloser (http://soap.genomics.org.cn/index.html), a 161,127 bp cp genome was obtained for C. heterophylla var. sutchuenensis. Annotation was performed using the Dual Organellar GenoMe Annotator (DOGMA) to generate a physical map of the cp genome (Wyman et al. Citation2004).
The circular cp genome of C. heterophylla var. sutchuenensis contains a pair of inverted repeat (IRa and IRb) regions, each of 26,615 bp, a large single-copy (LSC) region of 89,119 bp, and a small single-copy (SSC) region of 18,778 bp. It comprises 112 genes, including 78 protein-coding genes, 4 ribosomal RNA genes (16S, 23S, 5S, 4.5S), and 30 transfer RNA genes. Among the annotated genes, 17 protein-coding genes contain introns, including 15 with a single intron each and two with two introns each (clpP and ycf3). Interestingly, there is a trans-splicing gene (rps12) whose 5′ exon is located in the LSC and 3′ exon is located in the IR region.
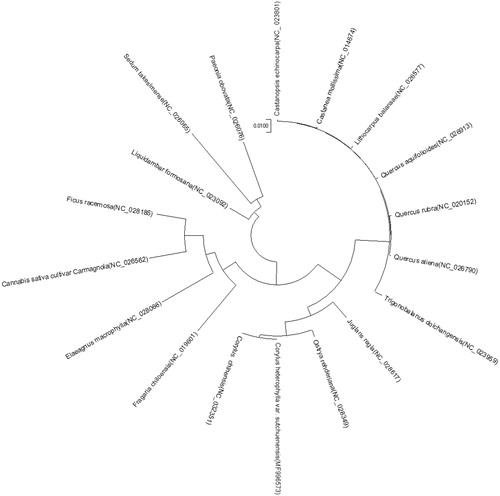
Maximum-likelihood phylogenetic analysis was performed based on the cp genomes of 11 Fagales plants and seven other outgroup plants using the MEGA6 software (Tamura et al. Citation2013). The cp genome of C. heterophylla var. sutchuenensis was shown to be similar to that of Corylus chinensis and closely related to that of Ostrya rehderiana of the family Betulaceae (). This complete cp genome can be used for subsequent population and cp genetic engineering studies, and especially to determine the phylogenetic position of C. heterophylla var. sutchuenensis in Corylus.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Bassil N, Boccacci P, Botta R, Postman J, Mehlenbacher S. 2013. Nuclear and chloroplast microsatellite markers to assess genetic diversity and evolution in hazelnut species, hybrids and cultivars. Genet Resour Crop Ev. 60:543–568. English.
- Boccacci P, Botta R. 2009. Investigating the origin of hazelnut (Corylus avellana L.) cultivars using chloroplast microsatellites. Genet Resour Crop Ev. 56:851–859.
- Erdogan V, Mehlenbacher S. 2000. Phylogenetic relationships of Corylus species (Betulaceae) based on nuclear ribosomal DNA ITS region and chloroplast matK gene sequences. Syst Bot 25:727–737.
- Hu GL, Cheng LL, Lan YP, Cao QC, Huang WG. 2017. The complete chloroplast genome sequence of Corylus chinensis Franch. Conserv Genet Resour. 9:119–121.
- Hu X. 1955. Handbook of economic plants. Beijing: Science Press.
- Huo H, Ma Q, Li J, Zhao T, Wang G. 2016. Study on the distribution of Corylus L. in China and the climatic evaluation of the suitable areas. J Plant Genet Res. 05:801–808.
- Li JL, Wang S, Jing Y, Wang L, Zhou S. 2013. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 48:72–78.
- Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Pan Q, Liu YJ, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Martins S, Simoes F, Mendonca D, Matos J, Silva AP, Carnide V. 2013. Chloroplast SSR genetic diversity indicates a refuge for Corylus avellana in northern Portugal. Genet Resour Crop Ev. 60:1289–1295. English.
- Palme AE, Vendramin GG. 2002. Chloroplast DNA variation, postglacial recolonization and hybridization in hazel, Corylus avellana. Mol Ecol. 11:1769–1780.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhang Y, Liu L, Liang W, Zhang Y. 2005. Chestnut hazelnut volume. Chinese fruit records. Beijing: Chinese Forestry Press.