Abstract
In this study, we first sequenced and described the complete mitochondrial genome and phylogeny of Dicrurus hottentottus. The whole genome of D. hottentottus was 16,963 bp in length, and contained 13 protein-coding genes, 22 transfer RNA genes, 2 ribosome RNA genes, and 1 non-coding control regions. The overall base composition of the mitochondrial DNA was 32.56% for A, 25.56% for T, 27.72% for C, and 14.15% for G, with a GC content of 41.87%. A phylogenetic tree strongly supported that D. hottentottus (Dicruridae) is closely related to Corvidae and Laniidae with a high probability.
Keywords:
Hair-crested Drongo (Dicrurus hottentottus Linnaeus, 1766) which belongs to the family Dicruridae of order Passeriformes, is mainly found in subtropical forests or tropical moist lowlands at an altitude between 0 and 1850 m. Dicrurus hottentottus has a very large range, extending from the south to southeast of Asia (BirdLife International Citation2019). The population size has not been quantified, but it is not believed to approach the thresholds for Vulnerable under the population size criterion and listed as Least Concern (LC) on the IUCN Red List (BirdLife International Citation2019; IUCN Citation2019). Pasquet et al. (Citation2007) addressed the phylogenetic relationships of the drongos (Dicruridae) at the species-level using sequences from two nuclear and two mitochondrial loci. Dicrurus hottentottus, though a commonly known species, has very few studies on its complete mitochondrial genome. Therefore, we sequenced the complete mitochondrial genome of D. hottentottus to enhance our understanding of the phylogeny of Dicruridae.
The specimen was collected from the town of Nabang in Yingjiang County, which is located northwestern of Yunnan Province in China, and stored at Faculty of Biodiversity Conservation, Southwest Forestry University. The total mitochondrial DNA was extracted from the muscle tissue using Next Generation Sequencing. The complete mitochondrial genome of D. hottentottus was submitted to the NCBI database under the accession number MK814795. Phylogenetic tree of the relationships among Corvidae, Laniidae, Dicruridae and Muscicapidae were presented using 15 species using maximum-likelihood (ML) methods available on the CIPRES Science Gateway v3.3 (Miller et al. Citation2010). Bayesian inference was calculated with MrBayes3.1.2 with a general time reversible (GTR) model of DNA substitution and a gamma distribution rate variation across sites (Ronquist and Huelsenbeck Citation2003). Sequences of Oriolidea (Oriolus chinensis) obtained from GenBank (NC_020424) were used as outgroups to root trees following Pasquet et al. (Citation2007) and Irestedt et al. (Citation2002).
The complete mitochondrial genome of D. hottentottus was 16,963 bp in length. A total of 38 mitochondrial genes were identified, including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and 1 non-coding control region (D-loop). Among these genes, nad6 and 8 tRNAs (trnQ, trnA, trnN, trnC, trnY, trnS2, trnP, and trnE) were located on the light strand (L-strand), while all of the remaining genes were located on the heavy strand (H-strand). The overall base composition of D. hottentottus mitogenome was 32.56% of A, 25.56% of T, 27.72% of C, and 14.15% of G, A + T content is 58.13%, which is higher than G + C content of 41.87%, similar to other Passeriformes (Yang et al. Citation2016; Liu et al. Citation2018).
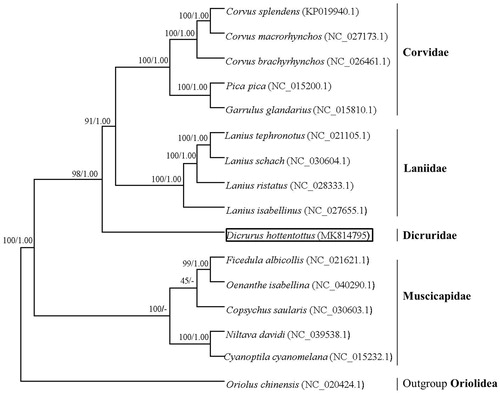
The reconstructed phylogenetic tree supported the placement of D. hottentottus in Dicruridae (). In our results, 15 species were clustered into four groups. Dicrurus hottentottus was closely related to Corvidae and Laniidae, and was strongly supported by the analyses of protein-coding genes. Thus, the mitochondrial genome reported here would be useful in the current understanding of the phylogeny and evolution of Dicruridae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- BirdLife International. 2019. Species factsheet: Indicator xanthonotus; [accessed 20 April 2019]. http://www.birdlife.org.
- Irestedt M, Fjeldsa J, Johansson US, Ericson P. 2002. Systematic relationships and biogeography of the Tracheophone suboscines (Aves: Passeriformes). Mol Phylogen Evol. 23:499–512.
- IUCN. 2019. The IUCN Red List of Threatened Species. Version 2019-1; [accessed 20 April 2019]. www.incnredlist.org.
- Liu R, Chen R, Liu J, Xiong Y, Kan X. 2018. Complete mitochondrial genome of Urocissa erythroryncha (Passeriformes: Corvidae). Mitochondrial DNA Part B. 3:691–692.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE); Nov 14; New Orleans. New Orleans, LA: Institute of Electrical and Electronics Engineers (IEEE).
- Pasquet E, Pons JM, Fuchs J, Cruaud C, Bretagnolle V. 2007. Evolutionary history and biogeography of the drongos (Dicruridae), a tropical Old World clade of corvoid passerines. Mol Phylogenet Evol. 45:158–167.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Yang MX, Wang QX, Xiao H, Yang C. 2016. Sequencing complete mitochondrial genome of Lanius sphenocercus sphenocercus (Passeriformes: Laniidae) using Illunima HiSeq 2500. Mitochondrial DNA Part B. 1:306–307.