Abstract
Rheum rhabarbarum is widely used in China to treat gout and constipation. We assembled and annotated the complete chloroplast genome of R. rhabarbarum. The length of the chloroplast genome is 161,723 bp, including a large single-copy (LSC) region of 87,004 bp, a small single-copy (SSC) region of 12,761 bp, and a pair of inverted repeat (IR) regions of 30,979 bp. The chloroplast genome included 130 genes, consisting of 36 tRNA genes, 8 rRNA genes, 2 pseudogenes, and 84 protein-coding genes. Phylogenetic analysis confirmed the position of R. rhabarbarum within the family Polygonaceae.
Keywords:
The Polygonaceae family comprises about 1200 species, distributed into about 48 genera. Rheum is a genus of about 60 species in the family Polygonaceae, widely distributed in Asia. Rheum rhabarbarum is naturally distributed in southern Siberia and north and central China (Wang et al. Citation2005). The Radix et Rhizoma of R. rhabarbarum can be used for the medicinal purpose (Liu et al. Citation2014). So far, no complete chloroplast (cp) genome of R. rhabarbarum has been published. Therefore, we presented the cp genome of this species in this study. The result could provide vital phylogenetic and genetic information for future studies in Polygonaceae.
Dried radix and Radix et Rhizoma powder of R. rhabarbarum was collected from National Institutes for Food and Drug Control, which batch number was 121676-201201. The voucher specimens were deposited at Tianjin State Key Laboratory of Modern Chinese Medicine. The total genomic DNA was isolated using Extract Genomic DNA Kit. Then, the Illumina Hiseq X Ten platform was used to sequence the DNA. The paired-end reads were assembled using NOVOPlasty 2.6.3 (Dierckxsens et al. Citation2017) with kmer 39. Seven contigs were mapped to reference (KR816224) to construct the cp genome. DOGMA (Wyman et al. Citation2004) and GeSeq (Tillich et al. Citation2017) were employed to annotate the complete plastid genome. The tRNAscan-SE (Lowe and Chan Citation2016) was used to verify tRNA genes. Finally, the preliminary annotated cp genome was checked carefully by comparing with published species of genus Rheum (R. officinale-MH572012; R. tanguticum-MH572013; R. palmatum-KR816224). The structural features of the cp genome were exhibited using OGDRAW (Lohse et al. Citation2013). Finally, the complete cp genome was submitted to GenBank (MK805223).
The chloroplast genome of R. rhabarbarum is 161,723 bp in length, including an LSC region of 87,004 bp, an SSC region of 12,761 bp, and a pair of IRs regions of 30,979 bp. It contains a total of 130 genes, including 84 protein-coding genes, 8 rRNA genes, 2 pseudogenes, and 36 tRNA genes.
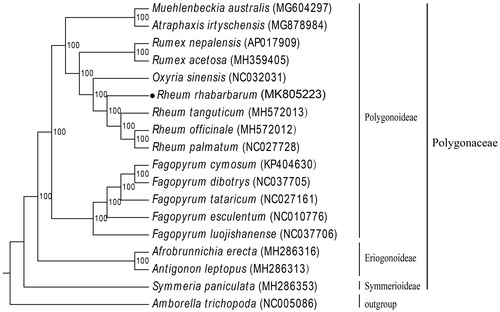
To help us understand the phylogenetic position of R. rhabarbarum in family Polygonaceae, R. rhabarbarum and other 16 Polygonaceae plastome sequences were used to reconstruct the phylogenetic tree. The complete cp genomes were aligned by MAFFT v7 (Katoh and Standley Citation2013). Then, RAxML v8.2.4 (Stamatakis Citation2014) was used to conduct the maximum likelihood tree (ML), using 1000 replicates of rapid bootstrap with the GTR + G substitution model (). As expected, R. rhabarbarum is in the same clade as other Rheum species. Moreover, the complete R. rhabarbarum cp genome can be used for further phylogenetic study within Polygonaceae.
Figure 1. ML phylogenetic tree of R. rhabarbarum with 17 species was constructed by chloroplast genome sequences. The numbers at each node indicate bootstrap support. GenBank accession numbers are given in brackets. Subfamilies and family of the sampled taxa are shown on the right. Amborellat richopoda was selected as outgroup.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Liu YN, Nan N, Lu BH, Xia WY, Wu XY, Bai QR, Gao J. 2014. First report of Phoma rhei as a pathogen of Rheum rhabarbarum in China. Plant Dis. 98:1154.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:575–581.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:54–57.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England). 30:1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Wang A, Yang M, Liu J. 2005. Molecular phylogeny, recent radiation and evolution of gross morphology of the rhubarb genus Rheum (Polygonaceae) inferred from chloroplast DNA trnL-F sequences. Ann Bot. 96:489–498.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics (Oxford, England). 20:3252–3255.
