Abstract
Flaveria bidentis (L.) Kuntze (Asteraceae) is one of the most hazardous invasive alien plant species and spread rapidly in China. In this study, the complete chloroplast genome of F. bidentis was sequenced using Illumina HiSeq X Ten platform. The results showed that the length of chloroplast genome of F. bidentis is 152, 230 bp, including a small single-copy region (18,362 bp), a large single-copy region (83,798 bp) and two inverted repeat regions (25,035 bp). The chloroplast genome contains 115 unique genes, including 81 protein-coding genes, four rRNA genes, and 30 tRNA genes. In total, we found 32 tandem repeats, 34 dispersed repeats and 36 SSRs in the chloroplast genome of F. bidentis. Phylogenetic analysis revealed that F. bidentis was in an independent clade and did not belong to the tribe Heliantheae Cass.
Flaveria bidentis (L.) Kuntze (Asteraceae) is an annual C4 plant and native plant species to South America (Wang et al. Citation2016). In 2001, F. bidentis was first found in Tianjin city in China and since then has been one of the most hazardous invasive weed species distributed in China. This invasive plant causes serious damages to local biodiversity and agricultural production (Zhang et al. Citation2010). In this study, the complete chloroplast (cp) genome of F. bidentis was sequenced and analyzed for the first time.
Fresh leave materials of F. bidentis were collected from Zhengzhou, Henan, China (N34°44′, E113°42′), and the voucher specimen (ZZU171011) was stored at the Herbarium of Zhengzhou University. The total DNA of F. bidentis was extracted from a single plant using the modified CTAB method and then purified by the Wizard DNA clean-up system (Promega Corp.) (Ref.). The DNA isolation was sequenced using the Illumina HiSeq X Ten platform at Novogene Biotech Co. (Beijing, China). The obtained raw data (6.2 G) was processed and assembled as described by Bakker et al. (Citation2016) and Wei et al. (Citation2017). The assembly was mapped to the reference Galinsoga quadriradiata cp genome sequence (Accession number NC_031853) using Geneious v10.1.3 software (Ma et al. Citation2013). The cp genome was annotated and checked manually in DOGMA (Wyman et al. Citation2004). The circular cp genome map was drawn in OGDRAW (Lohse et al. Citation2007). The tandem repeats, dispersed repeats, and Simple Sequence Repeat (SSRs) were detected using Tandem Repeats Finder (Kurtz et al. Citation2001), REPuter (Benson Citation1999), and MISA (Beier et al. Citation2017), respectively. The results showed that the cp genome of F. bidentis (NCBI accession number MK836182) exhibits typical circular structure and has a total length of 152,230 bp, with an LSC region of length 83,798 bp, an SSC region of 18,362 bp, and a pair of IR regions of 25,035 bp. The cp genome contains 115 unique genes, including 81 protein-coding genes, 30 tRNA, and four rRNA genes. Among annotated genes, 16 genes (10 protein-coding and six tRNA genes) contain one intron while two genes (clpP and ycf3) include two introns. The overall GC content of the cp genome is 37.66%. The 32 tandem repeats, 34 dispersed repeats, and 36 SSRs were identified in F. bidentis cp genome and can be used in genetic diversity research as molecular markers.
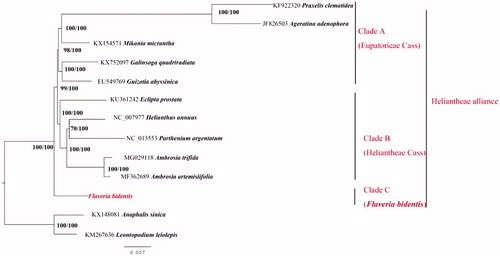
Ten cp genome sequences from different genera of Heliantheae alliance were used as ingroups to build Maximum Likelihood tree using RAxML-HPC2 (8.2.10) (Stamatakis Citation2014) and Maximum Parsimony tree using PAUP v4.0 (Swofford Citation2002). The phylogenetic trees showed three well-distinguished clades: clade A (Tribe Eupatoriea Cass.), clade B (Tribe Heliantheae Cass.) and clade C (F. bidentis) (). The results supported clade A + B as sister groups to clade C but separated F. bidentis from the tribe Heliantheae (Chen and Hind Citation2011). The phylogenetic analyses were in agreement with the previous study (Fu et al. Citation2016), which treated F. bidentis as an individual tribe.
Figure 1. Phylogenetic tree of 13 species based on the complete chloroplast genomes using the maximum-likelihood (RAXML) and Maximum parsimony (MP) methods. Anaphalis sinica and Leontopodium leiolepis were the outgroup species. The topology showed in this figure is from the ML tree. The numbers above the branches represent the RAXML/MP bootstrap values.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bakker FT, Lei D, Yu J, Mohammadin S, Wei Z, van de Kerke S, Gravendeel B, Nieuwenhuis M, Staats M, Alquezar-Planas DE, et al. 2016. Herbarium genomics: plastome sequence assembly from a range of herbarium specimens using an Iterative Organelle Genome Assembly pipeline. Biol J Linn Soc. 117:33–43.
- Beier S, Thiel T, Münch T, Scholz U, Mascher M. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics (Oxford, England). 33:2583–2585.
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580.
- Chen YS, Hind DJN. 2011. Heliantheae: Flora of China. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press. 852–878.
- Fu Z-X, Jiao B-H, Nie B, Zhang G-J, Gao T-G. 2016. A comprehensive generic-level phylogeny of the sunflower family: Implications for the systematics of Chinese Asteraceae. J Syst Evol. 54:416–437.
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29:4633–4642.
- Lohse M, Drechsel O, Bock R. 2007. Organellar Genome DRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genome. Curr Genet. 52:267–274.
- Ma C, Gunther S, Cooke B, Coppel RL. 2013. Geneious plugins for the access of PlasmoDB and PiroplasmaDB databases. Parasitol Int. 62:134–136.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30: 1312–1313.
- Swofford DL. 2002. PAUP. Phylogenetic analysis using parsimony (and other methods). Version 4. Mccarthy.
- Wang Y, Zhang YM, Li Q, Zhang FJ, Wan FH. 2016. Comparative study on the microbial community structure in different depth of rhizosphere soil in Flaveria bidentis invasion region. J Agric Univ Hebei. 39:35–42.
- Wei Z, Zhu SX, Van den Berg RG, Bakker FT, Schranz ME. 2017. Phylogenetic relationships within Lactuca L. (Asteraceae), including African species, based on chloroplast DNA sequence comparisons. Genet Resour Crop Evol. 64:55–71.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhang TR, HuangPu CH, Bai XM. 2010. Response of germinaton of different Flaveria bidentis populations to drought stress. Grassland and Turf. 30:79–83.
