Abstract
Fritillaria ussuriensis is the important herbal plant in the family Liliaceae, which is used in oriental medicine. In this study, we annotated and analyzed the complete chloroplast genome of F. ussuriensis. The chloroplast genome of F. ussuriensis has a circle structure with 151,478 bp in length and contains a pair of 26,296 bp inverted repeat (IR) regions, which were separated by a large single copy (LSC) region of 81,697 bp and a small single copy (SSC) region of 17,189 bp. The overall nucleotide composition of the complete chloroplast genome is as follows: A of 31.1%, T of 31.9%, C of 18.9%, and G of 18.1%, with a total G + C content of the chloroplast genome 37.0%. The complete chloroplast genome of F. ussuriensis encodes total 131 genes, including 85 protein-coding genes (PCGs), 38 transfer RNA genes (tRNAs), and eight ribosome RNA genes (rRNAs). The result of phylogenetic Maximum-Likelihood (ML) tree is set out that shows the closest relationship between F. ussuriensis and Fritillaria karelinii. The complete chloroplast genome provides basic data for further research of genus Fritillaria species useful genome resource in China.
Fritillaria ussuriensis is an Asian species of herbaceous plant in the Liliaceae family, which called Ping-bei-mu in Chinese (Day et al. Citation2014). The bulbs of Fritillaria ussuriensis are one of the traditional Chinese herbal medicines (Li et al. Citation2001). It has been used as the most important antitussive and expectorant drugs in China and other Asian countries for thousands years (Li et al. Citation2001). Fritillaria ussuriensis is distributed in temperate regions of the Northern hemisphere and in the lowland regions near sea level to 500 m of China, Russia, and Asian countries (Xing et al. Citation2000). Although the bulbs of the genus Fritillaria species have value in herbal medicine, different species are still used indiscriminately because of their morphological similarity and similar names, which consists of 140 known species (Xiang et al. Citation2016). At present, the genome information of F. ussuriensis was published too little. So, we annotated and analyzed the complete chloroplast genome of F. ussuriensis that can provide basic data for further research of genus Fritillaria species useful genome resource in China.
The specimen sample of Fritillaria ussuriensis was purchased from Jingyu town herbal market in Jilin (Jilin, Jingyu, China, 126.81E; 42.39N). The cpDNA of F. ussuriensis tissue was extracted using the CTAB method and stored the cpDNA of F. ussuriensis in Jilin Engineering Normal University Laboratory (Number JLENUL002). The cpDNA was purified and fragmented using the NEB Next UltraTM II DNA Library Prep Kit (NEB, BJ, and CN) that the cp genome was sequenced. The chloroplast genome of F. ussuriensis was assembled and annotated using MitoZ (Meng et al. Citation2019). The complete chloroplast genome map of F. ussuriensis was generated and drawn using OrganellarGenomeDRAW (Lohse et al. Citation2013).
The complete chloroplast genome of Fritillaria ussuriensis (NCBI No. MK6998921) is 151,478 bp in size that is a circle structure. It contains a quadripartite structure that consists of a large single-copy (LSC) region of 81,697 bp, a small single- copy (SSC) region of 17,189 bp, and a pair of inverted repeat (IR) regions of 26,296 bp in both. The overall nucleotide composition of the complete chloroplast genome is as follows: A of 31.1%, T of 31.9%, C of 18.9% and G of 18.1%, with a total G + C content of 37.0%. The chloroplast genome of F. ussuriensis encodes 131 genes, including 85 protein-coding genes (PCGs), 38 transfer RNA genes (tRNAs) and 8 ribosomal RNA genes (rRNAs). Seven PCGs (rpl2, rpl23, ycf2, ndhB, rps7, rps12 and ycf1), 8 tRNA genes (trnH-GUG, trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG and trnN-GUU) and 4 rRNA genes (rrn16S, rrn23S, rrn4.5S and rrn5S) are duplicated in both IR regions, which has 19 genes species in each IR.
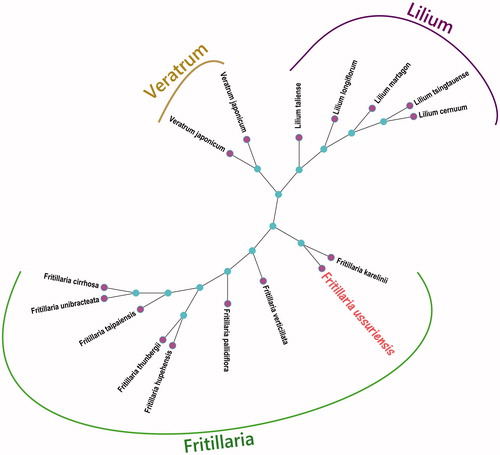
The phylogenetic tree was constructed based on 15 complete chloroplast genomes of the family Liliaceae plants with Fritillaria ussuriensis (), which were aligned using HomBlocks (Bi et al. Citation2018) and analyzed using Maximum-Likelihood (ML) methods. ML phylogenetic tree was represented using MEGA X (Kumar et al. Citation2018) and the bootstrap value based on 5,000 replicates is shown on each node, which edited using iTOL online web (https://itol.embl.de/). The results of the phylogenetic tree is set out in , which shows the complete chloroplast genome of Fritillaria ussuriensis is closest to the genus Fritillaria of Fritillaria karelinii (GenBank No. KX354691.1) in the genetic relationship. This study will provide basic data for further research of the genus Fritillaria species useful genome resource in China.
Figure 1. The phylogenetic tree constructed using Maximum-Likelihood (ML) analysis from 15 species complete chloroplast genomes of the family Liliaceae plants with Fritillaria ussuriensis. The bootstrap values were based on 5000 replicates. Fifteen family Liliaceae plants chloroplast genomes have been deposited in the GenBank and accession numbers are as follows: Fritillaria cirrhosa (KY646167.1), Fritillaria hupehensis (KF712486.1), Fritillaria karelinii (KX354691.1), Fritillaria pallidiflora (MG211822.1), Fritillaria taipaiensis (KC543997.1), Fritillaria thunbergii (KY646165.1), Fritillaria unibracteata (KF769142.1), Fritillaria verticillata (MG211823.1), Lilium cernuum (KX354692.1), Lilium longiflorum (KC968977.1), Lilium martagon (MF964219.1), Lilium taliense (KY009938.1), Lilium tsingtauense (KU230438.1), Veratrum japonicum (MG940972.1), and Veratrum patulum (KF437397.2).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22.
- Day PD, Berger M, Hill L, Fay MF, Leitch AR, Leitch IJ, Kelly LJ. 2014. Evolutionary relationships in the medicinally important genus Fritillaria L. (Liliaceae). Mol Phylogenet Evol. 80:11–19.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li S-L, Lin G, Chan S-W, Li P. 2001. Determination of the major isosteroidal alkaloids in bulbs of Fritillaria by high-performance liquid chromatography coupled with evaporative light scattering detection. J Chromatogr A. 909:207–214.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. doi:10.1093/nar/gkz173
- Xiang L, Su Y, Li X, Xue G, Wang Q, Shi J, Wang L, Chen S. 2016. Identification of Fritillariae bulbus from adulterants using ITS2 regions. Plant Gene. 7:42–49.
- Xing C, Mordak H, Tulipa L. 2000. In Flora China Flagellariaceae Marantaceae. Beijing (China): Science Press; p.123–136.
