Abstract
In this study, the complete mitochondrial genome (mitogenome) of Gracilaria spinulosa was reported for the first time. The complete mitogenome was 25,734 bp in length and contained 50 genes including 24 protein-coding, two rRNA, 23 tRNA genes, and one unidentified open reading frame (ORF). The structure of G. spinulosa mitogenome was similar to the other species in Gracilaria. The Bayesian phylogenetic analysis of Gracilariaceae revealed that G.spinulosa first clustered together with Gracilaria textorii, showing a distant relationship with Gracilaria chouae and Gracilaria vermiculophylla.
Gracilaria spinulosa (Okamura) Chang & B.-M.Xia (Gracilariaceae, Rhodophyta) is a flattened Gracilaria species with denticulate blade margins and is attached to rocky coral reefs or manmade concrete substrata in intertidal to subtidal zones (Zhang and Xia Citation1976; Lin Citation2006; Lin et al. Citation2012). Gracilaria spinulosa is a common marine macroalga occurring in Southeast Asian (Thailand, Vietnam, Philippines, Malaysia), Japan, southern China, Australia, and Caribbean sea, playing an important ecological role (Withell et al. Citation1994; Lin Citation2006; Yang et al. Citation2015). Lacking molecular data, the previous classification studies on G. spinulosa were mainly based on morphological features (Withell et al. Citation1994; Li et al. Citation2018). Here, we sequenced and annotated the complete mitogenome of G.spinulosa (GenBank accession number: MG592726) and constructed phylogenetic analysis to provide new molecular information.
Gracilaria spinulosa (specimen number: 2017060066) was collected from Yinggehai, Hainan Province in the southern China (18°30′36″N, 108°42′15″E) and deposited in the Culture Collection of Seaweed at the Ocean University of China. Total DNA was extracted by the modified CTAB method (Liu et al. Citation2017). Paired-end reads were sequenced using the HiSeq_Ten system (Illumina, USA). Gracilaria textori (GenBank accession number: NC_037892) was used as the reference sequence. The multiple protein and ribosomal RNA (rRNA) gene alignments were determined by BLAST similarity searches on the NCBI sequence database and compared to the published mitogenome of G. textori. Transfer RNA genes were identified using the tRNAscan-SE Search Server (Schattner et al. Citation2005).
The complete G. spinulosa mitogenome comprised a circular DNA molecule with the length of 25,734 bp. The base composition of the mitogenome is 33.88% A, 37.47% T, 14.17% G, and 14.48% C. The mitogenome contained 50 genes, including 24 protein-coding, two rRNA, 23 tRNA genes, and one unidentified open reading frame (ORF). The structure of G. spinulosa mitogenome was similar to the other species in Gracilaria. Of the 24 protein-coding genes, 22 (91.67%) terminated with the TAA stop codon, and two (8.33%) with TAG (nad3 and nad5). All protein-coding genes in mitogenome of G. spinulosa were found to have the start codon ATG. The length of 23 tRNA genes range from 72 to 93 bp with the typical clover leaf structure. The size of two rRNA genes was 2636 bp (rnl) and 1396 bp (rns), respectively.
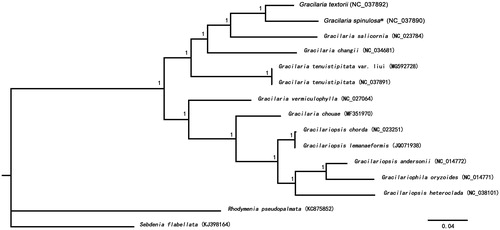
A total of 20 common protein-coding genes from the other 12 Gracilariaceae mitogenomes were used to compute a Bayesian phylogenetic tree using MrBayes version 3.1.2 software (Software Foundation, Inc., Cambridge, MA; Ronquist and Huelsenbeck Citation2003). Rhodymenia pseudopalmata (KC875852) and Sebdenia flabellata (KJ398164) was used as an outgroup. All algae were divided into two clades (). Phylogenetic analyses showed that G. spinulosa first clustered with Gracilaria textorii, showing a distant relationship with G. chouae and G. vermiculophylla. The complete mitogenome data provided in this work would help us to understand Gracilaria evolution.
Figure 1. Phylogenetic tree (Bayesian inference) based on 20 common protein-coding genes from 13 mitogenomes of Gracilariaceae, using Rhodymenia pseudopalmata (KC875852) and Sebdenia flabellata (KJ398164) as an outgroup. Support values for each node were calculated from Bayesian posterior probability (BPP). Asterisks following species names indicate newly determined mitogenomes.
References
- Li YM, Liu R, Yang N, Ding LP, Huang BX, Wang YX, Chen YW. 2018. Morphological taxonomy of four species of genus Gracilaria (Gracilariaceae, Rhodophyta) from Hainan Province, China. JTO. 37:29–37.
- Lin SM. 2006. Observations on flattened species of Gracilaria (Gracilariaceae, Rhodophyta) from Taiwan. J Appl Phycol. 18:671–678.
- Lin SM, Liu LC, Payri C. 2012. Characterization of Gracilaria vieillardii (Gracilariaceae, Rhodophyta) and molecular phylogeny of foliose species from the western Pacific Ocean, including a description of G. taiwanensis sp. nov. Phycologia. 51:421–431.
- Liu N, Wang GL, Li Y, Zhang L, Meinita MDN, Chen WZ, Liu T, Chi S. 2017. The complete mitochondrial genome of the economic red alga, Gracilaria chilensis. Mitochondr B Res. 2:716–717.
- Ronquist F, Huelsenbeck JP. 2003. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Withell AF, Millara AJK, Kraft GT. 1994. Taxonomic studies of the genus Gracilaria (Gracilariales, Rhodophyta) from Australia. Aust. Syst Bot. 7:281–352.
- Yang YF, Chai ZY, Wang Q, Chen WZ, He ZL, Jiang SJ. 2015. Cultivation of seaweed Gracilaria in Chinese coasta waters and its contribution to environmental improvements. Algal Res. 9:236–244.
- Zhang JF, Xia BM. 1976. Studies on Chinese species of Gracilaria. Stud Mar Sin. 11:91–166.
