Abstract
Castanopsis carlesii is one of the important evergreen broad-leaved forest species in the subtropical humid region of Eastern China and is distributed in south of the Yangtze River. We determined the complete chloroplast genome sequence for C. carlesii using Illumina sequencing data. The complete chloroplast sequence is 160,634 bp, include large single-copy (LSC) region of 90,318 bp, small single-copy (SSC) region of 18,960 bp, and a pair of invert repeats (IR) regions of 25,678 bp. Plastid genome contain 132 genes, 83 protein-coding genes, 40 tRNA genes, and eight rRNA genes. Phylogenetic analysis base on 26 chloroplast genomes indicates that C. carlesii is closely related to C. echinocarpa in Fagaceae.
Castanopsis carlesii belongs to Fagaceae; it is widely distributed in hills and mountains lower than 1300 m above sea level at the middle subtropical zone, and it is a kind of tree species with bias heat (He Citation2005). Subtropical evergreen broad-leaved forest is an oasis compared to the same latitude around the world, which is unique to China (Li et al. Citation2016). Castanopsis carlesii forest is one of the climax communities in the subtropical evergreen broad-leaved forests (Cai et al. Citation2005); it supports high levels of biomass pools in forest ecosystems. Therefore, this species is important to the material forest and shelter forest (Lin et al. Citation2017). The tree species has the advantages of fast growth, strong adaptability, barren resistance, and solid wood. It is an excellent timber tree species with light branches, small diameter wood, and bark, etc., and is also an excellent raw material for the cultivation of edible fungi (Liao et al. Citation1991). In this study, we report the complete chloroplast genome (cp) of C. carlesii based on Illumina pair-end sequencing data.
The plant material of C. carlesii was collected from Fujian province, China (Gushan Mountains, Fuzhou: 119°38′93.20″E, 26°05′24.03″N) and dried into silica gel immediately. The voucher specimen is kept at the Herbarium of College of Forestry, Fujian Agriculture and Forestry University (specimen code FAFU08013).
DNA extraction from fresh leaf tissue, with 500 bp randomly interrupted by the Covaris ultrasonic breaker for library construction. The constructed library was sequenced PE150 using Illumina Hiseq Xten platform, approximately 2GB data generated. Illumina data were filtered by script in the cluster (default parameter: −L 5, −p 0.5, −N 0.1). Complete plastid genome of C. echinocarpa (GeneBank accession: KJ001129) as reference, plastid genome of C. carlesii was assembled by GetOrganelle pipe-line (https://github.com/Kinggerm/GetOrganelle), it can get the plastid-like reads, and the reads were viewed and edited by Bandage (Wick et al. Citation2015). Assembled choroplast genome annotation base on comparison with C. echinocarpa using Geneious v 11.1.5 (Biomatters Ltd., Auckland, New Zealand) (Kearse et al. Citation2012). The annotation result was drawn with the online tool OGDRAW (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. Citation2013).
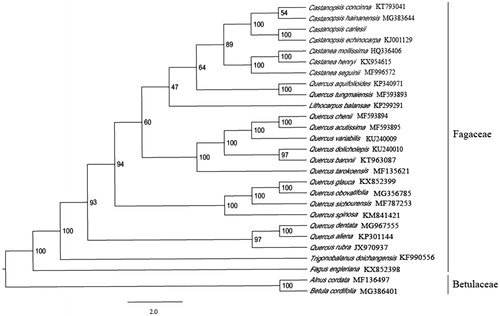
The complete plastid genome sequence of C. carlesii (GenBank accession: MK840978) was 160,634 bp in length, with a large single-copy (LSC) region of 90,318 bp, a small single-copy (SSC) region of 18,960 bp, and a pair of inverted repeats (IR) regions of 25,678 bp. Complete chloroplastid genome contains 132 genes; there were 83 protein-coding genes, 40 tRNA genes, and eight rRNA genes. The complete genome GC content was 36.8%. In order to reveal the phylogenetic position of C. carlesii with other members of Fagaceae, a phylogenetic analysis was performed based on 24 complete chloroplast genomes of Fagaceae, and two taxa (Alnus cordata, Betula cordifolia) as outgroups. They all downloaded from NCBI GenBank. The sequences were aligned using MAFFT v7.307 (Katoh and Standley Citation2013), and phylogenetic tree constructed using RAxML (Stamatakis Citation2014) The phylogenetic tree showed that C. carlesii was most closely related to C. echinocarpa with strong support ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cai XH, Zhong XH, Wang JX, He F, Liu XL, et al. 2005. Age-structure and dynamic of dominant population of Castanopsis carlesii community in Dujiangyan city, Sichuan, China. Wuhan Univ J Nat Sci. 4:694–700.
- He JY. 2005. Species diversity of Castanopsis carlesii community in Wuyishan Nature Reserve. J Xiamen Univ (Nat Sci). 44:7–10.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Li XJ, Liu XF, Xiong DC, Lin WS, Lin TW, Shi YW, Xie JS, Yang YS, et al. 2016. Impact of litterfall addition and exclusion on soil respiration in Cunninghamia lanceolate plantation and secondary Castanopsis carlesii forest in mid-subtropical China. Chinese J Plant Ecol. 40:447–457.
- Liao HZ, Zhang CN, Qiu DS, Chen ZZ, et al. 1991. Study on the production of Castanopsis carlesii plantation. J Fujian Coll Forester. 11:313–317.
- Lin KM, Lyu M, Jiang MH, Chen Y, Li YQ, Chen GS, Xie JS, Yang YS, et al. 2017. Improved allometric equations for estimating biomass of the three Castanopsis carlesii H. forest types in subtropical China. New Forests. 48:115–135.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31:3350–3352.