Abstract
Tulipa L. (Liliaceae) is among the world’s most beloved and economically important ornamental bulbs. Here, we reported the first chloroplast genome sequence of Tulipa. The plastome of T. altaica is 146,887 bp in length, including a large single-copy (LSC) of 78,433 bp, a small single-copy (SSC) of 16,628 bp, and a pair of inverted repeat regions (IRa and IRb) of 25,913 bp. It contains 111 unique genes consisting of 77 protein-coding genes, 30 tRNA genes, and four rRNA genes. The phylogenetic analysis revealed that Tulipa is sister to the clade formed by Erythronium and Amana.
Tulipa L. (Liliaceae), a spring-blooming perennial herbaceous genus, is native to central Asia, the Mediterranean, Europe, and North Africa and contains about 139 species in the world (Xing et al. Citation2017). It has been widely cultivated as ornamental plants worldwide for their various flower colors (Li et al. Citation2017). Despite its ecological and economic importance, there are a few molecular studies for breeding of this genus. Here, we sequenced and assembled the complete chloroplast genome sequence of T. altaica Pall. ex Spreng, which is the first one in the genus, and reconstructed the phylogenetic relationship with other Liliaceae species. Such a plastome sequence could provide abundant genetic information for identification, utilization, and breeding of Tulipa species.
Silica-gel-dried leaves were collected from one plant of Tulipa altaica in Xinjiang of China. The voucher specimen was deposited in the Herbarium of Zhejiang Univeristy (HZU). Genomic DNA was extracted using a standard CTAB method (Murray & Thompson Citation1980). Sequencing was conducted using HiSeq™2500 (Illumina, San Diego, California, USA). The raw data were trimmed and assembled into contigs using CLC Genomics Workbench v. 8.5.1 (http://www.clcbio.com). Then, all the resulting contigs were aligned to the reference chloroplast genome of Cardiocrinum cathayanum (Liliaceae, KX575836; Lu et al. Citation2017) using BLAST (http://blast.ncbi.nlm.nih.gov/), and the draft chloroplast genome was constructed by concatenating the resulting aligned contigs (≥90% similarity and query coverage) using Geneious v. 9.0.5 (http://www.geneious.com). The plastome was annotated using Geneious v. 9.0.5 by comparing with that of C. cathayanum, followed by manual inspection. The new annotated chloroplast genome sequence was deposited in GenBank (Accession No. MK673755).
The complete chloroplast genome of T. altaica was 146,887 bp long with a GC content of 37.1%. The genome consists of a pair of inverted repeat regions (IR) of 25,913 bp separated by a large single-copy (LSC) of 78,433 bp and a small single-copy (SSC) of 16,628 bp. Tulipa altaica encoded a total of 131 genes, of which 111 were unique and 20 were duplicated in the IR regions. Among the unique genes, there were 77 protein-coding genes (CDS), 30 transfer RNA (tRNA) genes, and four ribosomal RNA (rRNA) genes.
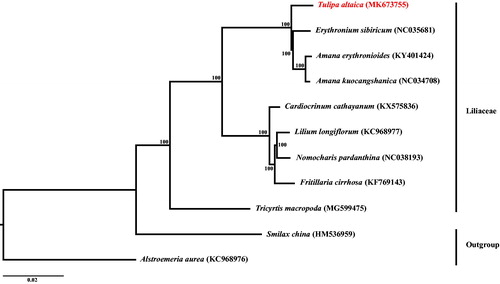
The phylogeny of Liliaceae was also reconstructed based on the multiple alignment of reported 11 chloroplast genomes with the Smilax china L. and Alstroemeria aurea Graham as outgroups using MAFFT v. 7.427 (Nakamura et al. Citation2018). The best-fitting model of nucleotide substitution was GTR + I + G, as determined by the Akaike Information Criterion (AIC) in jModelTest v. 2.1.7 (Darriba et al. Citation2012). ML (maximum likelihood) analysis was conducted using RAxML-HPC v. 8.2.8 with 1000 bootstrap replicates on the CIPRES Science Gateway website (Miller et al. Citation2010). Phylogenomic result () strongly supported that Tulipa is sister to the clade formed by Erythronium and Amana. The clade consisting of these three species is sister to the one including Cardiocrinum, Lilium, Nomocharis, and Fritillaria with 100% bootstrap values. This result was also congruent to that revealed in the previous studies based on cpDNA fragments (Kim and Kim Citation2018).
Disclosure statement
The authors are grateful to the uploaders for the open raw genome data in the public database. The authors report no conflicts of interest and are responsible for the content and writing of the paper.
Additional information
Funding
References
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.
- Kim JS, Kim JH. 2018. Updated molecular phylogenetic analysis, dating and biogeographical history of the lily family (Liliaceae: Liliales). Bot J Linnean Soc. 187:579–593.
- Li P, Lu RS, Xu WQ, Ohi-Toma T, Cai MQ, Qiu YX, Cameron KM, Fu CX. 2017. Comparative genomics and phylogenomics of East Asian Tulips (Amana, Liliaceae). Front Plant Sci. 8:1–12.
- Lu RS, Li P, Qiu YX. 2017. The complete chloroplast genomes of three Cardiocrinum (Liliaceae) species: comparative genomic and phylogenetic analyses. Front Plant Sci. 7:1–12.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE), New Orleans, LA, pp. 1–8.
- Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8:4321–4325.
- Nakamura T, Yamada KD, Tomii K, Katoh K. 2018. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 34:2490–2492.
- Xing GM, Qu LW, Zhang YQ, Xue L, Su JW, Lei JJ. 2017. Collection and evaluation of wild tulip (Tulipa spp.) resources in China. Genet Resour Crop Evol. 64:641–652.