Abstract
The accumulation of single nucleotide polymorphisms (SNPs) in the displacement loop (D-loop) of mitochondrial DNA (mtDNA) has been described in many cancers. In our previous studies, we have identified cancer risk and outcome associated SNPs in the D-loop of malignant fibrous histiocytoma (MFH). In the present study, we investigated the SNPs in the D-loop of mitochondria DNA (mtDNA) for their association with age-at-onset, metastasis, and relapse using a population-based series of MFH patients. Our data showed that the SNP site of nucleotide 16304 T/C was associated with age-at-onset using the Kaplan–Meier method and compared by the log-rank test. Meanwhile, allele 152C genotype was identified to be significantly associated with a high metastasis rate in MFH patients. In addition, patients with allele 16266C were proven to have a higher relapse rate compared with 16266T at a statistically significant level. Finally, we performed a multivariate analysis with the Cox proportional hazards model. The age-at-onset of patients with the minor allele C genotype was significantly earlier than that of patients with the T genotype at the 16304 site. In summary, the analysis of genetic polymorphisms in the D-loop may help to identify patient subgroups at a high risk of early onset, high metastasis and relapse rate in MFH, thereby helping to refine therapeutic decisions.
Introduction
The term malignant fibrous histiocytoma (MFH) was first coined by Ozzello et al. (Citation1963) and described by O’Brien and Stout (Citation1964). The next two decades confirmed and expanded the diagnostic entity of MFH. MFH is an aggressive soft tissue sarcoma which remains highly lethal malignancy and known to occur in various organs (Wanebo et al. Citation1992; Bairwa et al. Citation2017). MFH affects mostly the thighs and the trunk. Infrequently, it presents in the head and neck in adults (Gibbs et al. Citation2001). The lesions are usually diagnosed in high-grade stages and despite currently proposed therapies such as radiotherapy or chemotherapy, the patient’s prognosis is usually poor with a tendency to local recurrence and systemic metastasis (Weiss and Enzinger Citation1978). Early recognition is crucial to improve clinical outcome.
Mitochondrial DNA (mtDNA) carries out multiple cellular functions including ATP production, control of cell death, growth, development, integration of signals from mitochondria to nucleus and nucleus to mitochondria and various metabolic pathways. To perform its functions, mtDNA carries its own genome. Human mtDNA is 16 kb in length and is a closed-circular duplex molecule containing 37 genes, including 2 ribosomal RNAs (rRNAs), encoding 22 transfer RNAs (tRNAs), and 13 respiratory chain subunits that are essential for the respiratory functions of the mitochondria (Shadel and Clayton Citation1997). MtDNA is more prone to DNA damage and acquires mutations at a higher rate than nuclear DNA because of the high levels of reactive oxygen species (ROS), a lack of protective histones and a limited capacity for DNA repair in the mitochondria (DiMauro and Schon Citation2001; Conconi et al. Citation2005).
Somatic mutations and polymorphisms are located in an mtDNA non-coding region called the displacement loop (D-loop). In many cancers, sequence changes accumulate extensively in the mitochondrial D-loop region, which is significant for regulating the replication and expression of the mitochondrial genome, as it contains the leading-strand origin of replication and the main promoter for transcription (Taanman Citation1999; Nishikawa et al. Citation2001; Sanchez-Cespedes et al. Citation2001; Yoneyama et al. Citation2005). The D-loop contains a length of 1122 bp (nucleotide 16,024–16,569 and 1–576), according to the mitochondria database (http://www.mitomap.org). Single nucleotide polymorphisms (SNPs) in this region have been identified to be associated with disease risk and outcome of several cancers (Ding, Li, Wang, Fan, et al. Citation2012; Murtola et al. Citation2015; Thibodeau et al. Citation2015; Morgese et al. Citation2017). Previously, we identified that several SNPs, including 152T/C, 16390G/A, 16290C/T, 16304T/C, and AC deletion at sites 523 and 524 were associated with MFH risk and outcome. (Xun et al. Citation2016). The purpose of this study was to assess the correlations between germline SNPs in the D-loop and age-at-onset in MFH patients.
Methods
Tissue specimens and DNA extraction
According to the newly defined definition of MFH, we enrolled only pleomorphic sarcoma with no evidence of specific differentiation in our study. The histopathological specimens were reviewed by two pathologists, and if initial examination did not agree, the consensus was obtained after joint evaluation. Blood samples were collected from 80 MFH patients and 100 healthy controls. The genomic DNA was immediately extracted using the Wizard Genomic DNA extraction kit (Promega, Madison, WI, USA) and stored at −20 °C. All procedures were approved by the Human Tissue Research Committee in the hospital. All patients were provided with written informed consent before enrollment for the collection of samples and subsequent analysis.
PCR amplification and sequence analysis
The forward primer 5′-CCCCATGCTTACAAGCAAGT-3′ (nucleotide 16,190–16,209) and reverse primer 5′-GCTTTGAGGAGGTAAGCTAC-3′ (nucleotide 602–583) were used for amplification of a 982 bp product from mtDNA D-Loop region. PCR was performed according to the protocol of PCR Master Mix Kit (Promega, Madison, WI, USA) and purified prior to sequencing. Cycle sequencing was carried out with the Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystem, Foster City, CA, USA) and the products were then separated on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystem). Polymorphisms were confirmed by repeated analyses of both the strands.
Statistical analysis
We compared the patients age-at-onset and tumor characteristics among the different SNPs groups and related the results to the incidence of recurrence and metastasis. Differences between continuous variables were estimated by using the Mann–Whitney U test for comparing mean ranks. The survival curve was calculated using the Kaplan–Meier method and was compared by the log-rank test. Multivariate survival analysis was performed using a Cox proportional hazards model. All of the statistical analysis was done with the SPSS 18 software package (SPSS Company, Chicago, IL, USA). A p value of <.05 was considered statistically significant.
Results
We confirmed for the first time that SNPs in the D-loop were statistically associated with age-at-onset and clinical characteristics in MFH patients and some clinical characteristics also contribute to the early onset of MFH patients.
MtDNA genotype were associated with age-at-onset in MFH patients
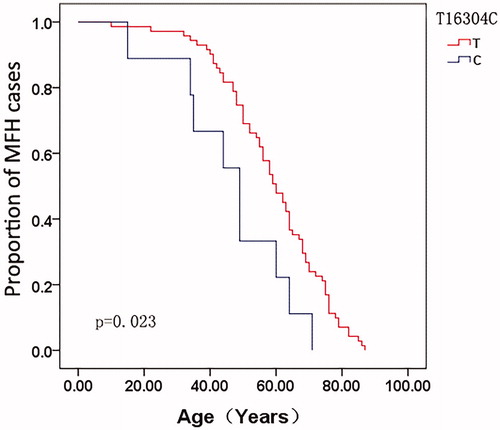
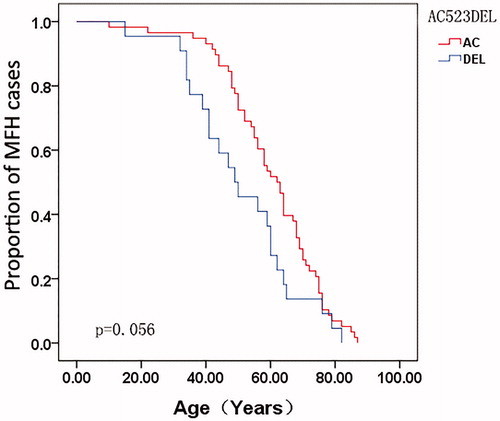
The correlations between the mtDNA genotype and age-at-onset were compared. A total of 80 MFH patients were enrolled in this study. For each SNP site, the age-at-onset curve was plotted using the Kaplan–Meier method for all MFH patients. The age-at-onset with the allele 16304T genotype was significantly earlier when compared with matched allele 16304C at the site 16304 in MFH patients (p = .023) (), while the allele AC del at site 523–524 was almost significantly associated with age-at-onset (p = .056) (), as revealed by the log-rank test.
MtDNA genotype was also associated with incidence of MFH metastasis and relapse
MtDNA genotype was also studied for their correlations with MFH metastasis and relapse. SNPs with a minor allele frequency of >5% were included; thus, the SNPs listed in were used for further analysis. The MFH patients were divided into two groups based on their genotype at each SNP site. Patients with the allele 152C were identified to be related to a high metastasis rate compared with allele 152T, which was statistically significant (p = .001) (). As for relapse, patients with the allele 16266C were proven to be significantly correlated with a high relapse rate compared with 16266T (p = .014) (), as revealed by the Mann–Whitney U test.
Table 1. Clinical characteristics and their association with age-at-onset age in MFH patients.
Table 2. The 27 SNP sites with a rare allele frequency higher than 5% and their associations with metastasis and relapse in MFH patients by Kaplan–Meier method.
Age-at-onset was associated with tumor location in lung in MFH patients
Age-at-onset was analyzed together with clinical characteristics, including tumor metastasis, relapse, tumor location, and pathological type, using the Kaplan–Meier method and then compared by log-rank test (). The tumor location was associated with age-at-onset at statistically significant levels (). The age-at-onset for patients with tumor location in the lung was significantly earlier when compared with other tumor locations in MFH patients (p < .001). Meanwhile, the association between age-at-onset and metastasis rate was almost statistically significant (p = .056).
Figure 3. Comparison of age-at-onset for MFH patients according to the tumor location by the Kaplan–Meier method.
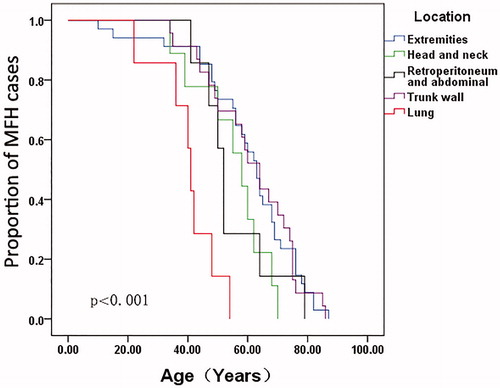
Table 3. Correlation of metastasis and relapse with other clinical characteristics in MFH patients.
We performed multivariate analysis with the Cox proportional hazards model including the factors of metastasis, tumor location, the SNP site 523–524 and the SNP site 16304. We coded patients with tumor metastasis as 1 and patients without metastasis as 0; patients with tumor location in the lung as 1 and patients with other tumor locations as 0; we coded C as 1 and T as 0 at 16304 allele site, AC del as 1 and AC as 0 at site 523–524. The age-at-onset of patients with allele 16304C genotype was significantly earlier compared with the allele 16304T genotype (relative risk, 2.146; 95% CI, 1.053–4.370; p = .035); the same was seen for patients who had tumor location in the lung than those with other tumor locations (relative risk, 7.495; 95% CI, 3.059–18.361; p<.001). These data demonstrated the strong independent prediction power of T16304C and tumor location (particularly in the lung) on age-at-onset for MFH patients.
Tumor relapse was associated with age and tumor location in MFH patients
As metastasis and relapse are the two main characteristics of MFH, their relationships with other clinical characteristics were also compared. The metastasis and relapse distribution of the MFH patients were listed in . Age, grade, location, and pathological type were not correlated with metastasis, while age and location were significantly associated with relapse in MFH patients.
Discussion
Mitochondria are central to cell metabolism, being the principal energy source of the cell, due to the cytochrome enzymes of terminal electron transport and the enzymes of the citric acid cycle, fatty acid oxidation, and oxidative phosphorylation.
Many SNPs in the mitochondrial D-loop region have been identified to be associated with cancer risk and outcome, including pancreatic cancer (Campa et al. Citation2018), breast cancer (Liu et al. Citation2015), prostate cancer (Freedman et al. Citation2018), esophageal squamous cell carcinoma (Zhang et al. Citation2010), lung cancer (Ding, Li, Wang, Jin, et al. Citation2012). MFH, a high-grade, undifferentiated pleomorphic sarcoma, is among the most malignant types of soft tissue sarcomas and exhibits repeated local recurrences, a high rate of metastasis, and poor prognoses (Matushansky et al. Citation2009). The pleomorphic form of MFH is associated with a 2-year survival rate of approximately 60% and a 42% rate of metastasis (Pobirci et al. Citation2011). We have previously identified that SNPs were significantly associated with MFH risk and post-operational survival (Xun et al. Citation2016).
However, age-at-onset, metastasis, and relapse associated SNPs in MFH patients have not been further studied. In this study, we identified SNPs in the D-loop were also predictors for age-at-onset of MFH patients. The allele of 16304C was associated with early onset of MFH compared with the allele 16304T with 9 MFH patients carried the C allele. Interestingly, it was one of the five SNPs which we previously identified for their association with post-operational survival.
Likewise, the SNP 152T/C was identified for its association with metastasis at a statistically significant level by using the Mann–Whitney U test (p = .001). In addition, we found the relapse rate of MFH patients was significantly related to mutation of C into T at 16266 loci (p = .014). Nevertheless, much larger sample size is also required to perform a comparison to see if there is any correlation for cancer risk at this site.
In our serial studies, we have identified that SNP site 152T/C was associated with MFH cancer risk as well as with MFH outcome (Xun et al. Citation2015, Citation2016). In other studies, SNP152 T/C was also correlated significantly with increased hepatocellular carcinoma risk (Chen et al. Citation2017). SNP sites 16304T/C and 523-524 del AC were associated not only with MFH outcome but also with MFH age-at-onset as well. Also, 16304 T/C was identified to have predictive power for disease outcome of human non-Hodgkin’s lymphoma (Fan et al. Citation2013; Diao et al. Citation2015). Whether these phenomena were just coincidences or these SNPs have some common functions in the process of cancer progression was unknown. Further studies were needed to elucidate the underlying mechanism of these SNPs.
We also analyzed the clinical characteristics for their associations with age-at-onset of the MFH patients. The tumor location and metastatic status of the MFH were also associated with age-at-onset. The age-at-onset for patients with tumor location in the lung was significantly earlier when compared with other tumor locations. According to literature, metastasis to the lung as secondary MFH is not uncommon. However, primary MFH of the lung is extremely rare, accounting for only less than 0.2% of lung neoplasms (Weiss and Enzinger Citation1978). Primary pulmonary MFH was a type of highly malignant sarcoma with poor prognosis (Lee et al. Citation2013). In our data, we collected seven primary pulmonary MFH patients, all of which have an early age onset than patients with any other tumor locations in our data. Contrary to our report, a previous study did not support any association between age-at-onset and tumor location in the lung. Small samples may be one of the reasons that led to research variation. However, it is of great value to explore the association between age-at-onset and tumor location in the lung, as well as in other organs in MFH patients.
In conclusion, we have proved for the first time that SNPs in the D-loop to be independent predictors for MFH age-at-onset as well as for metastasis and relapse. As a consequence, the analysis of genetic polymorphisms in the D-loop may help to identify patient subgroups at a high risk of early age-at-onset, high metastasis, and relapse rate in MFH, thereby helping to refine therapeutic decisions.
Disclosure statement
All authors declare that there is no competing interest.
Additional information
Funding
References
- Bairwa S, Sangwaiya A, Ansari M, Jindal A, Singla S, Yadav A. 2017. Malignant fibrous histiocytoma arising from renal capsule: an extremely rare entity. Indian J Pathol Microbiol. 60:402–404.
- Campa D, Pastore M, Capurso G, Hackert T, Di Leo M, Izbicki JR, Khaw KT, Gioffreda D, Kupcinskas J, Pasquali C. 2018. Do pancreatic cancer and chronic pancreatitis share the same genetic risk factors? A PANcreatic Disease ReseArch (PANDoRA) consortium investigation. Int J Cancer. 142:290–296.
- Chen C, Ba Y, Li D, Du X, Lia X, Yang H, An J, Xing J, Yang H, Dong G, et al. 2017. Genetic variations of mitochondrial genome modify risk and prognosis of hepatocellular carcinoma patients. Clin Res Hepatol Gastroenterol. 41:378–385.
- Conconi MT, Coppi PD, Liddo RD, Vigolo S, Zanon GF, Parnigotto PP, Nussdorfer GG. 2005. Tracheal matrices, obtained by a detergent-enzymatic method, support in vitro the adhesion of chondrocytes and tracheal epithelial cells. Transpl Int. 18:727–734.
- Diao L, Wei G, Su H, Li H, Song J, Gao Y, Guo Z. 2015. Sequence polymorphisms in the D-loop region of mitochondrial DNA and outcome of non-Hodgkin lymphoma. Mitochondrial DNA. 26:88–91.
- DiMauro S, Schon EA. 2001. Mitochondrial DNA mutations in human disease. Am J Med Genet. 106:18–26.
- Ding C, Li R, Wang P, Fan H, Guo Z. 2012. Sequence polymorphisms of the mitochondrial displacement loop and outcome of non-small cell lung cancer. Exp Thera Med. 3:861–864.
- Ding C, Li R, Wang P, Jin P, Li S, Guo Z. 2012. Identification of sequence polymorphisms in the D-loop region of mitochondrial DNA as a risk factor for lung cancer. Mitochondr DNA. 23:251–254.
- Fan H, Wang C, Guo Z. 2013. Single nucleotide polymorphisms in the mitochondrial displacement loop and age at onset of non-Hodgkin lymphoma. OncoTargets Ther. 6:1041–1045.
- Freedman JA, Wang Y, Li X, Liu H, Moorman PG, George DJ, Lee NH, Hyslop T, Wei Q, Patierno SR. 2018. Single-nucleotide polymorphisms of stemness genes predicted to regulate RNA splicing, microRNA and oncogenic signaling are associated with prostate cancer survival. Carcinogenesis. 39:879–888.
- Gibbs JF, Huang PP, Lee RJ, McGrath B, Brooks J, McKinley B, Driscoll D, Kraybill WG. 2001. Malignant fibrous histiocytoma: an institutional review. Cancer Invest. 19:23–27.
- Lee HJ, Yang HM, Choi YS, Park SH, Moon SH, Lee YS, Sung YC, Kim SJ. 2013. A therapeutic strategy for metastatic malignant fibrous histiocytoma through mesenchymal stromal cell-mediated TRAIL production. Ann Surg. 257:952–960.
- Liu M, Liu B, Song Y, Dong L. 2015. A rare case of carcinosarcoma of breast: Coexistence of mucinous carcinoma and malignant fibrous histiocytoma. J Cancer Res Therap. 11:1024
- Matushansky I, Charytonowicz E, Mills J, Siddiqi S, Hricik T, Cordon-Cardo C. 2009. MFH classification: differentiating undifferentiated pleomorphic sarcoma in the 21st Century. Expert Rev Anticancer Ther. 9:1135–1144.
- Morgese F, Soldato D, Pagliaretta S, Giampieri R, Brancorsini D, Torniai M, Rinaldi S, Savini A, Onofri A, Scarpelli M, et al. 2017. Impact of phosphoinositide-3-kinase and vitamin D3 nuclear receptor single-nucleotide polymorphisms on the outcome of malignant melanoma patients. Oncotarget. 8:75914–75923.
- Murtola TJ, Wahlfors T, Haring A, Taari K, Stenman UH, Tammela TL, Schleutker J, Auvinen A. 2015. Polymorphisms of genes involved in glucose and energy metabolic pathways and prostate cancer: interplay with metformin. Eur Urol. 68:1089–1097.
- Nishikawa M, Nishiguchi S, Shiomi S, Tamori A, Koh N, Takeda T, Kubo S, Hirohashi K, Kinoshita H, Sato E, et al. 2001. Somatic mutation of mitochondrial DNA in cancerous and noncancerous liver tissue in individuals with hepatocellular carcinoma. Cancer Res. 61:1843–1845.
- O'Brien JE, Stout AP. 1964. Malignant fibrous xanthomas. Cancer. 17:1445–1455.
- Ozzello L, Stout AP, Murray MR. 1963. Cultural characteristics of malignant histiocytomas and fibrous xanthomas. Cancer. 16:331–344.
- Pobirci DD, Bogdan F, Pobirci O, Petcu CA, Roşca E. 2011. Study of malignant fibrous histiocytoma: clinical, statistic and histopatological interrelation. Rom J Morphol Embryol = Revue Roumaine de Morphologie et Embryologie. 52:385–388.
- Sanchez-Cespedes M, Parrella P, Nomoto S, Cohen D, Xiao Y, Esteller M, Jeronimo C, Jordan RC, Nicol T, Koch WM, et al. 2001. Identification of a mononucleotide repeat as a major target for mitochondrial DNA alterations in human tumors. Cancer Res. 61:7015–7019.
- Shadel GS, Clayton DA. 1997. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 66:409–435.
- Taanman JW. 1999. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1410:103–123.
- Thibodeau SN, French AJ, McDonnell SK, Cheville J, Middha S, Tillmans L, Riska S, Baheti S, Larson MC, Fogarty Z, et al. 2015. Identification of candidate genes for prostate cancer-risk SNPs utilizing a normal prostate tissue eQTL data set. Nature Commun. 6:8653.
- Wanebo HJ, Koness RJ, MacFarlane JK, Eilber FR, Byers RM, Elias EG, Spiro RH. 1992. Head and neck sarcoma: report of the Head and Neck Sarcoma Registry. Society of Head and Neck Surgeons Committee on Research. Head Neck. 14:1–7.
- Weiss SW, Enzinger FM. 1978. Malignant fibrous histiocytoma: an analysis of 200 cases. Cancer. 41:2250–2266.
- Xun J, Li Z, Feng J, Gao S, Yang H, Song X. 2016. Single nucleotide polymorphisms in the mitochondrial displacement loop region and outcome of malignant fibrous histiocytoma. Mitochondr DNA A DNA Map Sequen Anal. 27:177–181.
- Xun J, Li Z, Song X, Wang X. 2015. Identification of sequence polymorphisms in the D-loop region of mitochondrial DNA as risk biomarkers for malignant fibrous histiocytoma. Mitochondr DNA. 26:380–383.
- Yoneyama H, Hara T, Kato Y, Yamori T, Matsuura ET, Koike K. 2005. Nucleotide sequence variation is frequent in the mitochondrial DNA displacement loop region of individual human tumor cells. Mol Cancer Res. 3:14–20.
- Zhang R, Wang R, Zhang F, Wu C, Fan H, Li Y, Wang C, Guo Z. 2010. Single nucleotide polymorphisms in the mitochondrial displacement loop and outcome of esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 29:155.