Abstract
Mitochondrial DNA (mtDNA) mutations play crucial roles in the pathogenesis and progression of human malignancies. However, studies reporting associations between mitochondrial DNA mutations and risks of esophageal squamous cell carcinomas (ESCC) have seldom been reported. In this study, we sequenced 22 cancer tissues and the corresponding adjacent non-cancerous tissues collected from 11 ESCC patients. The mtDNA sequence length ranged from 16,568–16,579 bp. The results indicated that there are several sensitive sites in the 22 mtDNAs, especially in the control region (CR) and protein-coding genes (PCGs). The 22 mtDNA sequences were classified into haplogroups B, D, G, M, and Z; haplogroup D and haplogroup M were shared within a wider distribution, but haplotype G was found only in two samples and all the patients showed the southern of East Asia or Northern East Asian haplogroups, the number and ratio of patients showed the southern of East Asia or Southeast Asia characters are equal. Our findings suggest that mtDNA haplogroups D and M might be potential risks for ESCC. In addition, our results suggest that mtDNA haplogroups and some sensitive sites confer genetic susceptibility to ESCC and they can serve as potential biomarkers for clinical diagnosis.
Introduction
The incidence of esophageal cancer (EC) is rapidly rising makes this cancer the fifth most common worldwide (O’Farrell et al. Citation2016). Nearly, 500,000 new cases of EC are diagnosed every year and with a poor overall five-year survival rate of just ∼14%, EC contributes to more than 400,000 deaths every year (Wang et al. Citation2018). There are two main types of EC, esophageal squamous cell carcinoma (ESCC) and adenocarcinoma with the predominant type in China being ESCC, accounting for 90% of the cases (Wang et al. Citation2018). While the mechanism(s) underlying ESCC carcinogens remains unclear, both environmental factors and genetics are thought to contribute with prior research highlighting the role of mtDNA (Beadnell et al. Citation2018; Wang et al. Citation2018).
The human mitochondrial genome is a small, circular double-stranded, relatively conserved DNA of 16,569 bp in length, containing 37 genes of 13 protein-coding genes (PCGs), 2 rRNA genes, and 22 tRNA genes. In addition, it also contains a variable sequence known as the control region (CR), which contains initiation sites for transcription and replication. Variations in mtDNA are very common and consequently have been widely used in forensic analyses, but nonetheless, mtDNA variations are increasingly recognized as important causes of human pathologies including cancers (Ding et al. Citation2012).
Previous studies have suggested that mtDNA mutations may be involved in the initiation of carcinogenesis and that such mutations may be potential biomarkers of carcinogenesis (Wallace Citation2005). It also indicated that some mtDNA haplogroups are associated with mitochondrial ROS production and play a role in cancer occurrence and development (Marcuello, Martínez-Redondo, Dahmani, Casajús, et al. Citation2009; Marcuello, Martínez-Redondo, Dahmani, Terreros, Citation2009; Martínez-Redondo et al. Citation2010; Zheng et al. Citation2012). However, very few studies have investigated the role of mtDNA variations in individual susceptibility to EC, especially to ESCC (Miyazono et al. Citation2002; Chatterjee et al. Citation2006; Loeb et al. Citation2008). In this study, we evaluated whether maternal haplogroups or mtDNA variations play crucial roles in Chinese patients with ESCC.
Materials and methods
Ethics
This study was approved by the institutional review board of Anhui Medical University with written informed consent obtained from all patients.
Tissue specimens
A total of 22 cancer tissues including the corresponding adjacent non-cancer tissues were collected from 11 ESCC patients who received treatment at the First Affiliated Hospital of Anhui Medical University, China, in 2016. All the samples were collected from patients of the mid-esophagus and all the cancer tissues were highly differentiated. Diagnoses of ESCC were histologically confirmed independently by two pathologists using hematoxylin-eosin (HE) stained sections. Samples for the study were collected immediately after the operation in 1.5 ml tubes and frozen in liquid nitrogen before storage at −80 °C in the Cancer Cell Biology Laboratory, School of Life Sciences, Anhui Medical University. The patients were numbered and the different tissues from the same patient were distinguished by adding the suffix, in detail, the ‘a’ refers to the primary ESCC tissue and the suffix ‘b’ refers to the adjacent non-cancerous tissue ().
Table 1. Summary of the 11 cases of ESCC used for sequencing analysis.
DNA amplification and sequencing
Genomic DNA from tissue was extracted using the Genomic DNA Isolation Kit (Sangon Biotech Co., Ltd. Shanghai, China). The mtDNAs were sequenced using next-generation sequencing technology by Shanghai Genesky Biotechnologies Inc. (Pudong District, Shanghai, CN). The 22 complete mtDNA sequences determined in this study were deposited in GenBank under accession numbers MK069558-MK069579.
Haplogroup classification, database comparison, and statistical analysis
The complete mtDNA sequence files were proofread and assembled in DNASTAR (DNAstar Inc., Madison, WI, USA). Multiple sequence alignments were analyzed by CLUSTAL_X.1.81 using the MEGA 6.0 software (Tamura et al. Citation2013). Sequence variations in complete mtDNA sequences were scored relative to the Cambridge Reference Sequence (rCRS) (Andrews et al. Citation1999). We classified the mtDNAs into specific haplogroups according to the updated East and Southeast Asian mtDNA phylogenetic trees, as well as the updated worldwide phylogenetic tree (http://www.phylotree.org). All the mtDNA sequences were compared in MITOMAP (MITOMAP: A Human Mitochondrial Genome Database. http://www.mitomap). The mtDNA haplotypes were classified into haplogroups according to the phylogenetic analyses of mtDNAs and Mitomap-Phylogeny. The phylogenetic trees were reconstructed with the ML and BI methods that were conducted in PAUP (version 4.0b8) and MrBayes 3.1.2 software (Strimmer et al. Citation1996), respectively.
Results and discussion
All the 22 mtDNA sequence length ranged from 16,568 to 16,579 bp (), the mean total nucleotide composition for the mtDNA sequences was: A, 30.9%; C, 31.3%; G, 13.1%; and T, 24.7%; the average AT content (55.6%) being slightly higher than the CG content (44.4%), which is similar to the rCRS () (Andrews et al. Citation1999). Mutations in the mtDNA may impact gene structure and cause transcriptional and translational defects with consequent dysfunction of the mitochondrial respiratory chain (Mohammed et al. Citation2015). A number of studies support an association between mtDNA mutations and cancers, particularly that certain mutations are correlated with various tumors (Fang et al. Citation2015). Researches indicated that 25–80% of mutations in mtDNA are found in cancers (Grzybowska-szatkowska and Slaska Citation2012). Variations occurred in CR (344 sites), PCGs (676 sites), tRNAs (60 sites), and rRNAs (103 sites) across all patients. Mutation rate in the CR (1.4%) is higher than that in PCGs (0.03%) in the patients with ESCC, suggesting that the mutations in these patients might affect the crucial mitochondrial function in the electron transport chain, which might, in turn, cause a high release of ROS and concomitant nuclear genome damage as well as cancer initiation and progression (Chen and Zhan Citation2009; Zheng et al. Citation2012). Comparing the cancer tissues and the corresponding adjacent non-cancer tissues from 11 ESCC patients in this study, the results indicate that the most significant variation is patient ID 28a v 28b (variation region length is 113 bp and the variation rate is 0.687%), the non-significant variation is patient ID 32a v 32b (variation region length is 4 bp and the variation rate is 0.024%) (). Human populations carry several mtDNA haplogroups defined by unique sets of mtDNA polymorphisms, reflecting mutations accumulated by a discrete maternal lineage (Riley et al. Citation2018). Previous studies have also demonstrated that some mtDNA haplogroups are associated with human diseases and influence longevity and carcinogens in conditions where mitochondrial ROS production is thought to play a role (Tanaka et al. Citation1998; Takagi et al. Citation2004; Wallace Citation2005; Cai et al. Citation2009). The association of mtDNA haplogroups with cancer has been extensively explored in breast cancer and prostate cancer (Cai et al. Citation2009; Salas et al. Citation2009 (Strimmer and von Haeseler, Citation1996) (Tamura et al., Citation2013); Fachal et al. Citation2014). Furthermore, several cancer-associated accumulations of mtDNA mutations have been reported, haplogroups A, C, Z, D, G, and N were observed to be enriched in lung cancer (Fang et al. Citation2015). Amongst the 22 mtDNA sequences screened, a total of 11 haplotypes were identified and classified into predicted haplogroups B, D, G, M, and Z (). Racial assignments of the haplogroups showed an equal number of patients segregated with East Asian/Southeast Asian or North East Asian haplogroups (). The phylogenetic relationships of all 22 mtDNA sequences are illustrated in . Of the 11 haplotypes, haplogroup B (18.2%) was shared in 03a and 03b, 14a and 14b; haplogroup D and haplogroup M were shared within a wider distribution (both are 27.3%); haplogroup D includes 07a, 07b, 81a, 81b, 32a, and 32b; while 05a, 57a, 05b, 31a, 28a, and 28b formed haplogroup M; haplogroup Z was found in samples 57b, 59a, 59b, and 31b, and was present 18.2%, haplotype G (9.1%) was found in two samples (70a and 70b). We found that the frequency of East Asian and Southeast Asian prevalent haplogroups (50.0%, 11/22) were equal to that of Northern East Asian prevalent haplogroups (50.0%, 11/22), the distributions of these haplogroups were different from those reported for lung cancer (Zheng et al. Citation2012; Fang et al. Citation2015). Our findings suggest that mtDNA D and M might be potential risk haplogroups for ESCC, which is similar to that of lung cancer (Ding et al. Citation2012; Zheng et al. Citation2012; Fang et al. Citation2015). In addition, the study reveals that mtDNA haplogroup D might be relatively more susceptible to DNA damage caused by the external ROS (Fang et al. Citation2015 ). Haplogroup D4 is one of the most characteristic mtDNA lineages found in Northeast Asian populations. It was reported that the 5178A of haplogroup D4 was enriched in Japanese centenarians, which is similar to the results of the previous study (Tanaka et al. Citation1998). However, because of the limit of samples, haplogroup G might decrease the risk of ESCC in the study. Our results suggest that mitochondrial DNA haplogroups and some sensitive sites confer genetic susceptibility to ESCC in patients and can serve as a potential biomarker for clinical diagnosis. However, due to the small sample size of this study, the association of mitochondrial DNA mutations with Chinese ESCC should be more deeply investigated in the future.
Figure 1. Distribution of variations in the 22 mitochondrial genomes sequenced. Traces show 13 PCGs combined, rRNA (12S rRNA and 16S rRNA combined), tRNA (all 22 tRNAs combined) and CR (all genes combined). X-axis is patient ID and Y-axis is number of base variations.
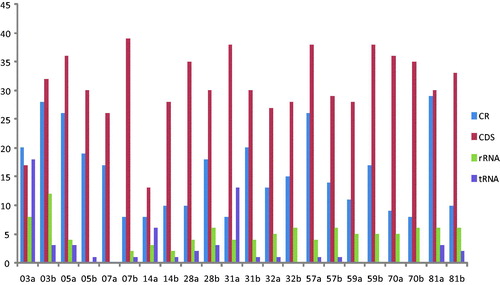
Figure 2. Phylogenetic relationships between the mtDNA sequences derived from 11 ESCC tissues and their corresponding adjacent non-cancerous tissues.
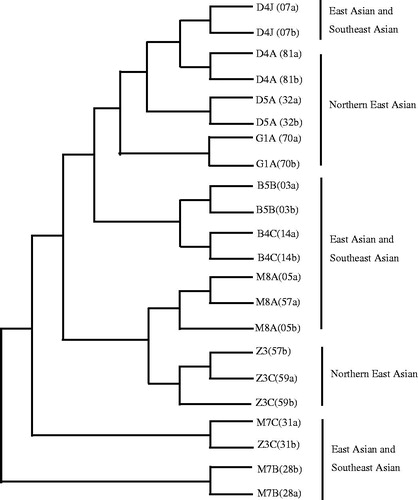
Table 2. Comparison of the cancer tissues and the corresponding adjacent non-cancer tissues from 11 ESCC patients in this study.
Table 3. Variable sites, mtDNA length and haplogroups of the 22 sequenced tissues.
Acknowledgments
We thank Qingyue Li, Chong Wang, and Caoling Xu for helping in the laboratory work and sequence analysis.
Disclosure statement
The authors declare no potential conflicts of interest.
Additional information
Funding
References
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. 1999. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 23:147.
- Beadnell TC, Scheid AD, Vivian CJ, Welch DR. 2018. Roles of the mitochondrial genetics in cancer metastasis: not to be ignored any longer. Cancer Metastasis Rev. 37:615–632.
- Cai X-y, Wang X-f, Li S-l, Qian J, Qian D-g, Chen F, Yang Y-j, Yuan Z-y, Xu J, Bai Y, et al. 2009. Association of mitochondrial DNA haplogroups with exceptional longevity in a Chinese population. PLoS ONE. 4:e6423.
- Chatterjee A, Mambo E, Sidransky D. 2006. Mitochondrial DNA mutations in human cancer. Oncogene. 25:4663–4674.
- Chen D, Zhan H. 2009. Study on the mutations in the D-loop region of mitochondrial DNA in cervical carcinoma. J Cancer Res Clin Oncol. 135:291–295.
- Ding CM, Li RJ, Wang P, Jin PL, Li SM, Guo ZJ. 2012. Identification of sequence polymorphisms in the D-loop region of mitochondrial DNA as a risk factor for lung cancer. Mitochondr DNA. 23:251–254.
- Fachal L, Gómez-Caamaño A, Álvarez Iglesias V, Gómez Carballa A, Calvo P, Salas A, Vega A. 2014. No association between typical European mitochondrial variation and prostate cancer risk in a Spanish cohort. J Human Genet. 59:411–414.
- Fang Y, Huang J, Zhang J, Wang J, Qiao F, Chen HM, Hong ZP. 2015. Detecting the somatic mutations spectrum of Chinese lung cancer by analyzing the whole mitochondrial DNA genomes. Mitochondr DNA. 26:56–60.
- Grzybowska-Szatkowska L, Slaska B. 2012. Polymorphisms in genes encoding mt-tRNA in female breast cancer in Poland. Mitochondr DNA. 23:106–111.
- Jin XJ, Zhang JJ, Gao YJ, Ding KY, Wang NS, Zhou D, Jen J, Cheng SJ. 2007. Relationship between mitochondrial DNA mutations and clinical characteristics in human lung cancer. Mitochondrion. 7:347–353.
- Loeb LA, Bielas JH, Beckman RA. 2008. Cancers exhibit a mutator phenotype: clinical implications. Cancer Res. 68:3551–3557.
- Marcuello A, Martínez-Redondo D, Dahmani Y, Casajús JA, Ruiz-Pesini E, Montoya J, López-Pérez MJ, Díez-Sánchez C. 2009. Human mitochondrial variants influence on oxygen consumption. Mitochondrion. 9:27–30.
- Marcuello A, Martínez-Redondo D, Dahmani Y, Terreros JL, Aragonés T, Casajús JA, Echavarri JM, Quílez J, Montoya J, López-Pérez MJ, et al. 2009. Steady exercise removes VO (2max) difference between mitochondrial genomic variants. Mitochondrion. 9:326–330.
- Martínez-Redondo D, Marcuello A, Casajús JA, Ara I, Dahmani Y, Montoya J, Ruiz-Pesini E, López-Pérez MJ, Díez-Sánchez C. 2010. Human mitochondrial haplogroup H: the highest VO2max consumer-is it a paradox? Mitochondrion. 10:102–107.
- Miyazono F, Schneider PM, Metzger R, Warnecke-Eberz U, Baldus SE, Dienes HP, Aikou T, Hoelscher AH. 2002. Mutations in the mitochondrial DNA D-Loop region occur frequently in adenocarcinoma in Barrett's esophagus. Oncogene. 21:3780–3783.
- Mohammed F, Rezaee Khorasany AR, Mosaieby E, Houshmand M. 2015. Mitochondrial A12308G alteration in tRNA(Leu(CUN)) in colorectal cancer samples. Diagn Pathol. 19:115.
- O’Farrell NJ, Feighery R, Picardo SL, Lynam-Lennon N, Biniecka M, McGarrigle SA, Phelan JJ, Mac Carthy F, O’Toole D, Fox EJ, et al. 2016. Changes in mitochondrial stability during the progression of the Barrett’s esophagus disease sequence. BMC Cancer. 16:497.
- Riley V, Erzurumluoglu AM, Rodriguez S, Bonilla C. 2018. Mitochondrial DNA haplogroups and breast cancer risk factors in the avon longitudinal study of parents and children (ALSPAC). Genes. 9(8):1.
- Salas A, Fachal L, Marcos-Alonso S, Vega A, Martinón-Torres F. 2009. Investigating the role of mitochondrial haplogroups in genetic predisposition to meningococcal disease. PLoS ONE. 4:e8347.
- Strimmer, K, von Haeseler, A. 1996. Quartet Puzzling: A Quartet Maximum-Likelihood Method for Reconstructing Tree Topologies. Molecular Biology and Evolution. 13:964–969. DOI:10.1093/oxfordjournals.molbev.a025664
- Takagi K, Yamada Y, Gong J-S, Sone T, Yokota M, Tanaka M. 2004. Association of a 5178C->A (Leu237Met) polymorphism in the mitochondrial DNA with a low prevalence of myocardial infarction in Japanese individuals. Atherosclerosis. 175:281–286.
- Tamura, K, Stecher, G, Peterson, D, Filipski, A, Kumar, S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution. 30:2725–2729. DOI:10.1093/molbev/mst197
- Tanaka M, Gong JS, Zhang J, Yoneda M, Yagi K. 1998. Mitochondrial genotype associated with longevity. Lancet. 351:185–186.
- Wallace DC. 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 39:359–407.
- Wang K, Chen DM, Meng Y, Xu JJ, Zhang QY. 2018. Clinical evaluation of 4 types of microRNA in serum as biomarkers of esophageal squamous cell carcinoma. Oncol Lett. 16:1196–1204.
- Zheng S, Qian P, Li F, Qian G, Wang C, Wu G, Li Q, Chen Y, Li J, Li H, et al. 2012. Association of mitochondrial DNA variations with lung cancer risk in a Han Chinese population from southwestern China. PLoS ONE. 7:e31322.
