Abstract
In this study, the complete mitochondrial genome of the social aphid Pseudoregma bambucicola was assembled and annotated. The genome spans 16,631 bp in length with a high A + T content (85.1%), containing 13 protein-coding genes, 22 tRNA genes, two rRNA genes, one control region locating between srRNA and tRNAIle, and a repeat region located between tRNAGlu and tRNAPhe. All the tRNAs except tRNACys and tRNASer(gct) exhibit the typical clover-leaf like structures. The lengths of lrRNA and srRNA are 1301 bp and 758 bp, respectively. The order of genes between COIII and tRNAGlu exhibits significant difference for P. bambucicola. A phylogenetic analysis for P. bambucicola and 22 other aphids based on 13 protein-coding genes is also presented.
The aphid Pseudoregma bambucicola (Hormaphidinae: Cerataphidini) is an important pest of Bambusa bamboos in subtropical areas. This species can produce polymorphic nymphs with behaviour differentiation and some first instar nymphs behave as soldiers to protect their clones (Sakata and Itô Citation1991). Pseudoregma bambucicola belongs to the tribe Cerataphidini, in which several aphid genera can produce soldiers. To date, in Hormaphidinae, only one complete mitochondrial genome of Hormaphis betulae from the tribe Hormaphidini has been reported (Li et al. Citation2017). Here, we report the complete mitochondrial genome of P. bambucicola, representing the first mitogenome of Cerataphidini.
Samples used for sequencing were collected in June 2017 in Xiamen of Fujian province, China. Voucher specimen (HL20170622-4) was deposited in the Insect Systematics and Diversity Lab at Fujian Agriculture and Forestry University, Fuzhou, China. The mitogenome was sequenced on an Illumina platform and assembled using NovoPlasty v. 2.7.1 (Dierckxsens et al. Citation2017). Then, we manually annotated the genome with the MITOS Webserver (Bernt et al. Citation2013). The sequence has been deposited in GenBank (accession number: MK847518).
The total length of P. bambucicola mitogenome is 16,631 bp, longer than majority of published aphid mitogenomes. The nucleotide composition is typically A + T biased (85.1%) with similar patterns also found in other aphids (Wang et al. Citation2015; Ren et al. Citation2016; Ren and Wen Citation2017; Chen et al. Citation2019; Hong et al. Citation2019; Voronova et al. Citation2019). The mitogenome contains 13 protein-coding genes (PCGs), 22 tRNA genes (tRNAs), two rRNA genes (rRNAs), and one control region. Fifteen tRNAs and nine PCGs are located on the forward strand (J-strand) while the remaining genes are transcribed on the reverse strand (N-strand). The mean length of tRNAs is 65 bp, ranging from 52 bp to 73 bp. All 22 tRNAs exhibit the typical clover-leaf like secondary structures except tRNACys and tRNASer(gct). For the two rRNAs, lrRNA (1301 bp) is located between tRNALeu(tag) and tRNAVal, and srRNA (758 bp) between tRNAVal and the control region. The control region (709 bp) with a high A + T content (95.1%) is located between srRNA and tRNAIle. In addition, a long repeat region between tRNAGlu and tRNAPhe also exists, which was thought unique to some aphid lineages (Wang et al. Citation2013, Citation2015). However, this region is absent in the H. betulae mitogenome (Li et al. Citation2017). Previous studies revealed that the gene order of most aphid mitogenomes, including H. betulae, are identical to that of inferred ancestral arrangement of insects (Wang et al. Citation2013). However, six genes (J-strand) between COIII and tRNAGlu have undergone significant rearrangements in P. bambucicola.
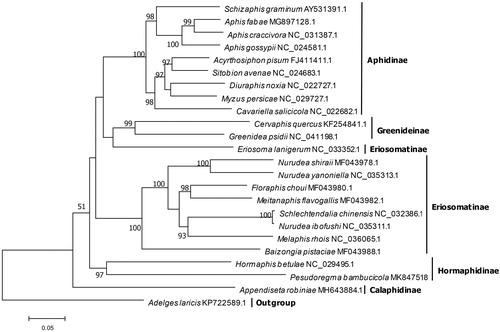
A phylogenetic analysis based on 13 protein-coding genes from P. bambucicola and 22 other aphids with complete mitogenomes was undertaken using Adelges laricis as outgroup (). A maximum-likelihood phylogenetic tree was reconstructed using IQ-TREE (Nguyen et al. Citation2015), which showed the two Hormaphidinae species, P. bambucicola and H. betulae, clustered together with strong support. However, some basal nodes with low supports masked the clear relationships between some subfamilies.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Chen J, Wang Y, Qin M, Jiang LY, Qiao GX. 2019. The mitochondrial genome of Greenidea psidii van der Goot (Hemiptera: Aphididae: Greenideinae) and comparisons with other Aphididae aphids. Int J Biol Macromol. 122:824–832.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18
- Hong B, Zhang F, Hu ZQ, Zhao HY. 2019. The complete mitochondrial genome of Indomegoura indica (Hemiptera: Aphididae). Mitochond DNA B Res. 4:882–883.
- Li YQ, Chen J, Qiao GX. 2017. Complete mitochondrial genome of the aphid Hormaphis betulae (Mordvilko) (Hemiptera: Aphididae: Hormaphidinae). Mitochondrial DNA A DNA Mapp Seq Anal. 28:265–266.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Ren ZM, Bai X, Harris AJ, Wen J. 2016. Complete mitochondrial genome of the Rhus gall aphid Schlechtendalia chinensis (Hemiptera: Aphididae: Eriosomatinae). Mitochond DNA B Res. 1:849–850.
- Ren ZM, Wen J. 2017. Complete mitochondrial genome of the North American Rhus gall aphid Melaphis rhois (Hemiptera: Aphididae: Eriosomatinae). Mitochond DNA B Res. 2:169–170.
- Sakata K, Itô Y. 1991. Life history characteristics and behaviour of the bamboo aphid, Pseudoregma bambucicola (Hemiptera: Pemphigidae), having sterile soldiers. Insect Soc. 38:317–326.
- Voronova NV, Warner D, Shulinski R, Levykina S, Bandarenka Y, Zhorov D. 2019. The largest aphid mitochondrial genome found in invasive species Therioaphis tenera (Aizenberg, 1956). Mitochond DNA B Res. 4:730–731.
- Wang Y, Chen J, Jiang LY, Qiao GX. 2015. Hemipteran mitochondrial genomes: features, structures and implications for phylogeny. Int J Mol Sci. 16:12382–12404.
- Wang Y, Huang XL, Qiao GX. 2013. Comparative analysis of mitochondrial genomes of five aphid species (Hemiptera: Aphididae) and phylogenetic implications. PLoS ONE. 8:e77511