Abstract
Laodelphax striatellus (Fallén, 1826) is one of key rice pests in Northeast Asia. We have determined the mitochondrial genome of L. striatellus collected in a mid-western part of Korean peninsula. The circular mitogenome of L. striatellus is 16,359 bp long including 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNAs, and a single large non-coding region of 1,972 bp. The base composition was AT-biased (77.3%). In comparison of the two Chinese L. striatellus mitogenomes with Korean mitogenome, total 140 and 40 single nucleotide polymorphisms and 166 and 118 insertions and deletions were identified, presenting intra-species variations based on geographical distribution.
Laodelphax striatellus (Fallén, 1826) (small brown planthopper; SBPH), of which body size is less than 4 mm, is a hemimetabolous insect widely distributed in Asia, Europe, and Northern Africa (Wilson and Claridge Citation1991). High density of SBPH could cause severe economic damage on rice, Oryza sativa L., by directly sucking and indirectly transmitting viral pathogens, including rice stripe virus and rice streaked dwarf virus, with piercing mouthparts (i.e. stylets; Hibino Citation1996; Seo et al. Citation2016). Two mitogenomes (Song and Liang Citation2009; Zhang et al. Citation2013) and one whole genome (Zhu et al. Citation2017) originated from China (Jiansu province and Beijing) were published, insufficient to uncover geographical differences among SBPH populations in Northeast Asia, where SBPHs can overwinter and migrate from China to Korea and Japan by westerlies in early summer (Otuka et al. Citation2012).
Here we first mitochondrial genome of Korean SBPH collected in Suwon, South Korea in 1970 (37°16′20″N, 126°57′57″E; voucher was deposited in InfoBoss Cyber Herbarium (IN; Seo BY, INH-00018; South Korea). DNA was extracted using CTAB-based DNA extraction method manually (iNtRON biotechnology, Inc., Korea). Raw sequences obtained from Illumina NextSeq500 (Macrogen Inc., South Korea) were filtered using Trimmomatic 0.33 (Bolger et al. Citation2014) and de novo assembled using Velvet 1.2.10 (Zerbino and Birney Citation2008) and MITOBim 1.9.1 (Hahn et al. Citation2013) and gaps including 548-bp AT-rich region were closed with SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17, and SAMtools 1.9 (Li et al. Citation2009; Li Citation2013) as well as PCR, respectively. Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate mitochondrial genome based on that of Chinese L. striatellus (NC_013706; Zhang et al. Citation2013).
Laodelphax striatellus mitochondrial genome (GenBank accession is MK838101) is 16,359 bp, which is the shortest among three SBPH mitogenomes. Its nucleotide composition is AT-biased (A + T is 77.3%) and contains 13 protein-coding genes, two rRNAs, and 22 tRNAs. The control region, presumably corresponding to single largest non-coding AT-rich region (1,972 bp, A + T is 83.8%), is shorter than those of Chinese SBPH mitogenomes (2,040 bp in NC_013706 and 2,042 bp in JX880068).
Gene order of three SBPH mitogenomes is identical. In comparison of two Chinese L. striatellus mitogenomes (NC_013706 and JX880068) with Korean SBPH, 140 and 40 single nucleotide polymorphisms and 166 and 118 insertions and deletions were identified, respectively, presenting intra-species variations based on geographical distribution, similar to Nilaparvata lugens (Choi et al. Citation2019; Park, Kwon, et al. Citation2019) and Chilo suppresallis (Park, Xi, et al. Citation2019).
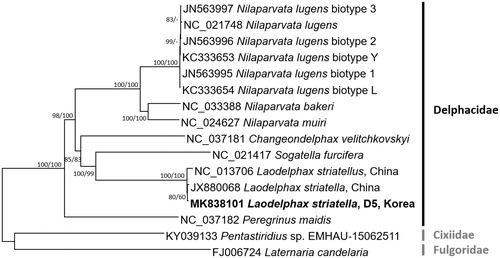
We inferred the phylogenetic relationship of 16 mitogenomes, seven species of Delphacidae including three L. striatellus and two outgroup species (Pentastiridius sp. and Laternaria candelaria). Multiple sequence alignment was conducted using MAFFT 7.388 (Katoh and Standley Citation2013). Neighbour-joining (10,000 bootstrap repeats) and maximum-likelihood (1,000 bootstrap repeats) phylogenetic trees were constructed using MEGA X (Kumar et al. Citation2018). Seven species in Delphacidae are monophyletic and three L. striatellus mitogenomes are more divergent than six N. lugnes mitogenomes (). This mitochondrial genome will be helpful in understanding geographical intraspecies variations of L. striatellus.
Figure 1. Neighbour-joining (10,000 bootstrap repeats) and maximum-likelihood (1,000 bootstrap repeats) phylogenetic trees of seven insect species in the family Delphacidae, three Laodelphax striatellus (MK838101 in this study, NC_013706, and JX880068), six Nilaparvata lugens (NC_021748, JN563997, JN563996, JN563995, KC333653, and KC333654), Nilaparvata bakeri (NC_033388), Nilaparvata muiri (NC_024627), Sogatella furicefera (NC_021417), Changeondelphax velitchkovskyi (NC_037181), and Peregrinus maidis (NC_037182), and as outgroup species, Pentastiridius sp. (KY039133) and Laternaria Candelaria (FJ006724). Phylogenetic tree was drawn based on neighbour-joining tree. The numbers above branches indicate bootstrap support values of neighbour-joining and maximum-likelihood phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Choi NJ, Lee B-C, Park J, Park J. 2019. The complete mitochondrial genome of Nilaparvata lugens (Stål, 1854) captured in China (Hemiptera: Delphacidae): investigation of intraspecies variations between countries. Mitochondrial DNA B. 4:1677–1678.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129–e129.
- Hibino H. 1996. Biology and epidemiology of rice viruses. Annu Rev Phytopathol. 34:249–274.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv. 13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Otuka A, Zhou Y, Lee G-S, Matsumura M, Zhu Y, Park H-H, Liu Z, Sanada-Morimura S. 2012. Prediction of overseas migration of the small brown planthopper, Laodelphax striatellus (Hemiptera: Delphacidae) in East Asia. Appl Entomol Zool. 47:379–388.
- Park J, Kwon W, Park J, Kim H-J, Lee B-C, Kim Y, Choi NJ. 2019. The complete mitochondrial genome of Nilaparvata lugens (stål, 1854) captured in Korea (Hemiptera: Delphacidae). Mitochondrial DNA B. 4:1674–1676.
- Park J, Xi H, Kwon W, Park C-G, Lee W. 2019. The complete mitochondrial genome sequence of Korean Chilo suppressalis (Walker, 1863) (Lepidoptera: Crambidae). Mitochondrial DNA B. 4:850–851.
- Seo B, Kwon Y, Jung J, Kim G. 2016. Feeding behavior of the small brown Planthopper, Laodelphax striatellus (Hemiptera: Delphacidae) on rice plants based on EPG waveform, honeydew excretion, and microsection analysis. Korean J Appl Entomol. 55:351–358.
- Song N, Liang A-P. 2009. Complete mitochondrial genome of the small brown planthopper, Laodelphax striatellus (Delphacidae: Hemiptera), with a novel gene order. Zool Sci. 26:851–860.
- Wilson MR, Claridge MF. 1991. Handbook for the identification of leafhoppers and planthoppers of rice. Wallingford: CAB International.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhang K-J, Zhu W-C, Rong X, Zhang Y-K, Ding X-L, Liu J, Chen D-S, Du Y, Hong X-Y. 2013. The complete mitochondrial genomes of two rice planthoppers, Nilaparvata lugens and Laodelphax striatellus: conserved genome rearrangement in Delphacidae and discovery of new characteristics of atp8 and tRNA genes. BMC Genomics. 14:417.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
- Zhu J, Jiang F, Wang X, Yang P, Bao Y, Zhao W, Wang W, Lu H, Wang Q, Cui N. 2017. Genome sequence of the small brown planthopper Laodelphax striatellus. GigaScience. 6:1–12.
