Abstract
In this study, the complete mitochondrial genome of a male individual of Murina cyclotis from Guangxi province, China, was sequenced and analyzed. The genome is a circular molecule of 16,463 bp length, containing 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and a control region. Most of the genes were encoded on the H-strand, except for 8 tRNA and ND6 genes. Phylogenetic trees of the complete mitochondrial genome were constructed using RAxML. Our result suggests that M. cyclotis is closely related to M. leucogaster from Korea. The complete mitochondrial genome sequence of M. cyclotis will be helpful for future taxonomic and phylogenetic studies on Murina.
The species diversity in genus Murina is high in Asian region, with 39 recognized species. The current number of species listed is twice the number of species listed until 2005 (Csorba and Bates Citation2005; Kuo et al. Citation2006, Citation2009; Csorba et al. Citation2007; Kruskop and Eger Citation2008; Furey et al. Citation2009; Csorba et al. Citation2011; Eger and Lim Citation2011; Francis and Eger Citation2012; Ruedi et al. Citation2012; Soisook, Karapan, Satasook, Thong, et al. Citation2013; Soisook, Karapan, Satasook, Bates Citation2013; He et al. Citation2015; Chen et al. Citation2017). Among these species, Murina cyclotis is a medium-sized species. Apparently, this species is similar to M. huttoni externally (Francis and Eger Citation2012; Liu and Wu Citation2019). However, phylogenetic evidences such as mitochondrial genes (e.g., COX1, Cyt b and ND1) do not support a close relationship between them (Eger and Lim Citation2011). Surprisingly, the barcoding investigation further indicated occurrences of cryptic species within this taxon (Francis and Eger Citation2012; Soisook, Karapan, Satasook, Thong, et al. Citation2013).
In this study, complete mitochondrial genome (MK747248) of a male individual M. cyclotis (GZHU 16335) collected from Diding country, Guangxi Province (23°06′51.6″N 105°58′12.6″E), China, was sequenced. Complete genomic DNA was extracted from liver tissue using MiniBEST Universal Genomic DNA Extraction Kit (TAKARA, Dalian) and was sequenced using Illumina Hiseq X.
The total length of the mitochondrial genome is 16,463 bp. It encodes 13 protein-coding genes, two rRNA genes, 22 tRNA genes, and a control region, the same pattern as in the other three Murina mitochondrial genomes (Yoon et al. Citation2013; Yoon and Park Citation2016; Zhang et al. Citation2016). All the protein-coding genes (total 11,399 bp) were encoded in the H-strand except for ND6 in L-strand. Among the 13 protein-coding genes, most common start codon is ATG while the ND2 and ND5 start with ATA, ND3 starts with ATT. Seven protein-coding genes stop with TAA, only Cyt b ends with AGA. The rest of five genes stop with an incomplete termination codons TA– (ND1, ND3) and T–– (ND2, COX3, ND4), which may be modified by the polyadenylation after transcription (Ojala et al. Citation1981). Total length of the 22 tRNA genes is 1505 bp ranging from 62 bp (tRNA-Ser) to 75 bp (tRNA-Leu) and can be folded into typical cloverleaf secondary structure except for the tRNA-Ser (AGC). The control region with 1018 bp in length is located between the tRNA-Pro and tRNA-Phe genes, and it has high simple repeat sequence GCAATC.
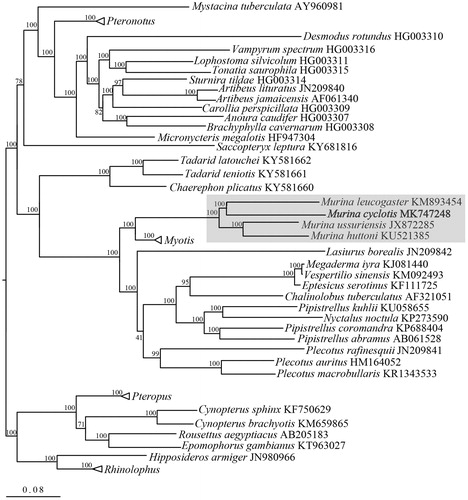
The phylogenetic relationships of M. cyclotis were inferred using mitochondrial 13 protein-coding genes of 89 complete mitochondrial genome sequences. PartitionFinder 1.1.1 (Lanfear et al. Citation2012) was used to select the best partitioning scheme and regional best-fit models of nucleotide evolution. In the phylogenetic tree, all Murina species were clustered together, and it revealed closer relationship of M. cyclotis with M. leucogaster rather than with M. huttoni. Besides, Murina and Myotis clustered more closely together and clearly separated from other genera in Vespertilionidae (). Such patterns suggest morphogroups within Murina (e.g. ‘suilla-group’ and ‘cyclotis-group’) do not represent phylogenetic assemblages. Given the fact that a multi-lineages and cryptic species were observed within M. cyclotis using DNA barcoding technique (Francis and Eger Citation2012), future taxonomic and mitochondrial genome sequences throughout its distribution range are desired. Here, the first mitochondrial genome of M. cyclotis is provided, and it can benefit future phylogenetic and evolutionary studies on this genus and taxon.
Figure 1. Molecular phylogenetic analysis of 89 species of order Chiroptera (Pteronotus contains 4 species, Myotis contains 33 species, Pteropus contains 4 species, Rhinolophus contains 11 species) based on complete mitochondrial genome. The phylogenetic tree was constructed using maximum likelihood method. Percentages of trees where associated taxa were clustered together were shown next to branches, and Murina clade highlighted in shade.
Acknowledgements
We thank the help of supervisors and teammates.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen J, Liu T, Deng H, Xiao N, Zhou J. 2017. A new species of Murina bats was discovered in Guizhou Province, China. Cave Res. 2:1–10.
- Csorba G, Bates PJJ. 2005. Description of a new species of Murina from Cambodia (Chiroptera: Vespertilionidae: Murininae). Acta Chiropterol. 7:1–7.
- Csorba G, Son NT, Saveng I, Furey NM. 2011. Revealing cryptic bat diversity: three new Murina and redescription of Murina tubinanris from Southeast Asia. J Mammal. 92:891–904.
- Csorba G, Thong DV, Bates PJJ, Furey NM. 2007. Description of a new species of Murina from Vietnam (Chiroptera: Vespertilionidae: Murininae). Occasional Papers. Lubbock, TX: Museum of Texas Tech University. Vol. 268, p. 1–12.
- Eger JL, Lim BK. 2011. Three new species of Murina from Southern China. Acta Chiropterol. 13:227–243.
- Francis CM, Eger JL. 2012. A review of tube-nosed bats (Murina) from Laos with a description of two new species. Acta Chiropterol. 14:15–38.
- Furey NM, Thong VD, Bates PJJ, Csorba G. 2009. Description of a new species belonging to the Murina ‘suilla-group’ (Chiroptera: Vespertilionidae: Murininae) from north Vietnam. Acta Chiropterol. 11:225–236.
- He F, Xiao N, Zhou J. 2015. A new species of Murina from China (Chiroptera: Vespertilionidae). Cave Res. 2:1–6.
- Kruskop SV, Eger JL. 2008. A new species of tube-nosed bat Murina (Vespertilionidae, Chiroptera) from Vietnam. Acta Chiropterol. 10:213–220.
- Kuo HC, Fang YP, Csorba G, Lee LL. 2006. The definition of Harpiola (Vespertilionidae: Murininae) and the description of a new species from Taiwan. Acta Chiropterol. 8:11–19.
- Kuo HC, Fang YP, Csorba G, Lee LL. 2009. Three new species of Murina (Chiroptera: Vespertilionidae) from Taiwan. J Mammal. 90:980–991.
- Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701.
- Liu SY, Wu Y. 2019. Handbook of the mammals of China. Fuzhou: The Straits Publishing & Distributing Group Press.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Ruedi M, Biswas J, Csorba G. 2012. Bats from the wet: two new species of Tube-nosed bats (Chiroptera: Vespertilionidae) from Meghalaya, India. Revue Suisse Zool. 119:111–135.
- Soisook P, Karapan S, Satasook C, Bates PJJ. 2013. A new species of Murina (Mammalia: Chiroptera: Vespertilionidae) from peninsular Thailand. Zootaxa. 3746:567–579.
- Soisook P, Karapan S, Satasook C, Thong VD, Khan FAA, Maryanto I, Gsorba G, Furey N, Aul B, Bates PJJ. 2013. A review of the Murina cyclotis complex (Chiroptera: Vespertilionidae) with descriptions of a new species and subspecies. Acta Chiropterol. 12:271–292.
- Yoon KB, Kim HR, Kim JY, Jeon SH, Park YC. 2013. The complete mitochondrial genome of the Ussurian tube-nosed bat Murina ussuriensis (Chiroptera: Vespertilionidae) in Korea. Mitochondrial DNA. 24:397–399.
- Yoon GB, Park YC. 2016. The complete mitogenome of the Korean greater tube-nosed bat, Murina leucogaster (Chiroptera: Vespertilionidae). Mitochondrial DNA Part A. 27:2145–2146.
- Zhang QP, Cong HY, Yu WH, Kong LM, Wang YY, Li YC, Wu Y. 2016. The mitochondrial genome of Murina huttoni rubella (Chiroptera: Vespertilionidae) from China. Mitochondrial DNA Part B. 1:438–440.
