Abstract
Strawberry (Fragaria x ananassa) is an economically and important fruit species that is grown extensively around the entire world. In this study, we obtained the complete chloroplast genome of F. x ananassa. The chloroplast genome of F. x ananassa is 155,529 bp in size, containing a large single-copy region (85,523 bp), a small single-copy region (18,136 bp) and a pair of IR regions (each one of 25,935 bp). The overall GC contents of the chloroplast genome is 37.2%. The chloroplast genome of F. x ananassa contains 135 genes, including 90 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The phylogenetic maximum-likelihood (ML) tree was constructed and based on 18 species plants and F. x ananassa chloroplast genomes, which F. x ananassa is closest related to Fragaria vesca subsp. vesca in phylogenetic relationship. This complete chloroplast genome can be further used for genomic studies, phylogenetic analyses, and genetic engineering studies of the genus Fragaria.
Strawberry (Fragaria x ananassa) is the economic berry of the family Rosaceae and an allopolyploid species with diverse, which originated in the 1700s from a natural hybridization between two strawberries (Folta and Gardiner Citation2009). Strawberry originated from South America and widely cultivated in Europe, America, Asia, and other countries at present that is planted more than 20,000 varieties all over the world. Fragaria x ananassa is one of the favourite fruits and widely consumed throughout the world with the pleasant aroma, flavour, taste, and texture. Simultaneously, it is rich vitamins, minerals, antioxidant compounds and thus considered, which receives more and more attention from growers and consumers (Forbes-Hernandez et al. Citation2016; Patrick et al. Citation2019). In order to study the genus Fragaria and family Rosaceae species genetic evolution and genetic population, we choose to study the chloroplast genome of strawberry (F. x ananassa) as a representative. So, we obtained and researched the complete chloroplast genome of F. x ananassa, which contributes to study genomic and genetic engineering of the genus Fragaria and family Rosaceae in the future.
The specimen sample of F. x ananassa was isolated and purchased from Strawberry picking garden in Putian district of Fujian province (Fujian, China, 119.08E; 25.52N). The total genomic DNA of F. x ananassa was extracted using the optimized CTAB method and stored in Putian University (No. PTU002). The total genomic DNA was used for the shotgun library construction and sequenced using the Illumina HiSeq 4000 Sequencing Platform System ((Illumina Co., San Diego, CA). Quality control was performed to remove low-quality reads and adapters using the FastQC (Andrews Citation2015). The chloroplast genome was assembled using the Plann (Huang and Cronk Citation2015) and annotated using the DOGMA (Wyman et al. Citation2004). The tRNA genes were further identified using tRNAscan-SE v2.0 (Lowe and Chan Citation2016). The physical map of the chloroplast genome of F. x ananassa was generated using OrganellarGenomeDRAW (Lohse et al. Citation2013).
The complete chloroplast genome of F. x ananassa (GenBank accession No.MK8043121) was a circle molecular genome with the length of 155,529 bp in size, which was containing a large single-copy region (LSC) of 85,523 bp, a small single- copy region (SSC) of 18,136 bp and a pair of inverted repeat regions (IRs) of 25,935 bp in each one. The overall nucleotide composition is 31.1% of A, 31.7% of T, 18.9% of C, and 18.3% of G, with the total GC content is 37.2%. The CP genome of F. x ananassa comprised 135 genes, including 90 protein-coding genes (PCGs), 37 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. In the IR regions, 20 genes were found duplicated in each one, which includes nine PCG species (rpl2, rpl23, ycf2, ycf15, ndhB, rps7, rps12, ycf68, and ycf1), seven tRNA species (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU), and four rRNA species (rrn16, rrn23, rrn4.5, and rrn5).
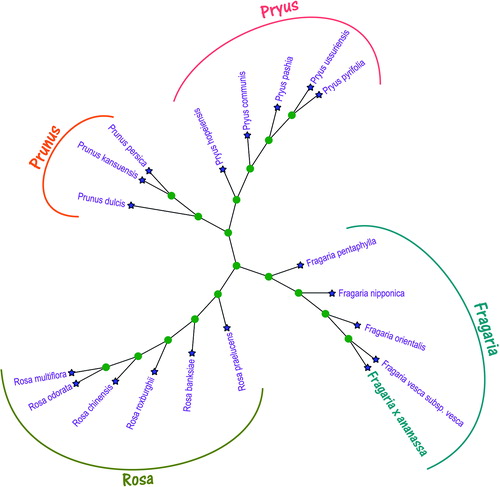
The chloroplast genomes of 18 species plants in the family Rosaceae and F. x ananassa were used for constructing phylogenic maximum-likelihood (ML) trees and analyzed the phylogenetic relationship with them. The phylogenetic tree was reconstructed using maximum-likelihood (ML) methods and performed using the RaxML v8 (Stamatakis Citation2014), which the bootstrap value was calculated using 5000 replicates to assess node support and all the nodes were inferred with strong support by the ML methods. The evolutionary tree was constructed using the MEGA X (Kumar et al. Citation2018) and edited using the iTOL v4 (Ivica and Peer Citation2019). The phylogenetic tree results () showed that F. x ananassa is located in the genus Fragaria and is clustered and closest related to Fragaria vesca subsp. Vesca (NC_015206.1). The complete chloroplast of Strawberry can be significance for genomic studies and genetic engineering studies of the genus Fragaria and family Rosaceae.
Figure 1. Maximum-likelihood (ML) phylogenetic tree of Fragaria x ananassa with 18 other species plants in the family Rosaceae based on the complete chloroplast genome sequences. GenBank accession numbers of complete chloroplast genome sequences are listed as follows: Fragaria nipponica NC_035500.1, Fragaria orientalis NC_035501.1, Fragaria pentaphylla NC_034347.1, Fragaria vesca subsp. vesca NC_015206.1, Prunus dulcis KY085904.1, Prunus kansuensis KF990036.1, Prunus persica HQ336405.1, Pryus communis KX450884.1, Pryus hopeiensis MF521826.1, Pryus pashia NC_034909.1, Pryus pyrifolia NC_015996.1, Pryus ussuriensis KX450882.1, Rosa banksiae MK361034.1, Rosa chinensis NC_038102.1, Rosa multiflora MG893867.1, Rosa odorata KF753637.1, Rosa praelucens MG450565.1, and Rosa roxburghii NC_032038.1.
Acknowledgements
Jing Huang thanks Dr Shoukai Lin and Prof. Jincheng Wu for their kind suggestions.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. Available online at http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Folta KM, Gardiner SE. 2009. Genetics and genomics of Rosaceae, vol. 6. New York: Springer; p. 411–506.
- Forbes-Hernandez TY, Gasparrini M, Afrin S, Bompadre S, Mezzetti B, Quiles JL, Giampieri F, Battino M. 2016. The healthy effects of strawberry polyphenols: which strategy behind antioxidant capacity? Crit Rev Food Sci Nutr. 56:S46–S59.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Ivica L, Peer B. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW–a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:54–57.
- Patrick PE, Thomas JP, Robert V, Michael AH, Marivi C, Michael RM, Ronald DS, Scott JT, Andrew DLN, Ching MW, et. al. 2019. Origin and evolution of the octoploid strawberry genome. Nat Genet. 51:541–547.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
