Abstract
The plant genus Melastoma of the family Melastomataceae is comprised of nine species and one variety in China. Melastoma dodecandrum is the only creeping species of this genus. Previous study has reported the complete chloroplast genome of M. dodecandrum from Guangzhou, China, but there may be some differences between plant populations from different regions. Herein, we reported the complete chloroplast genome of M. dodecandrum from Fuzhou, China, which was assembled from Pacbio and whole genome data was sequenced. The sequence has a circular molecular length of 156,598 bp and contained 129 genes. Phylogenetic analysis indicated that M. dodecandrum was closely related to M. candidum in Melastomataceae. The study aims to provide insights for the future studies on the differences in molecular evolution level between plant populations of M. dodecandrum and taxonomy of Melastoma.
The genus Melastoma has undergone species radiation. It might have happened during the last one million years (Renner and Meyer Citation2001), it has approximately 100 species in total with nine species and one variety in China (Chen Citation2007). Melastoma dodecandrum, the only creeping species within Melastoma genus, is widely distributed in tropical and subtropical regions of China except Hainan (Editors Commitment Citation1984). Recent study (Zheng et al. Citation2019) has finished the complete chloroplast genome sequencing of M. dodecandrum (GenBank accession MH748092, the sequence was not released on NCBI, accessed on 11 May 2019), which is a circular molecule of 156,611 bp in length from Guangzhou, China. Herein, we report a different result of 156,598 bp length in M. dodecandrum from Fuzhou, Fujian, to integrate the future studies on the differences in molecular evolution level between plant populations from different regions of M. dodecandrum and taxonomy of Melastoma.
For the re-assembly of this chloroplast genome, the sequence data were extracted from the total sequencing data from whole-genome of a M. dodecandrum individual sampled from Fuzhou, Fujian, China (N 26°13′19″, E 119°17′27″) using Pacbio sequencing effort. The voucher specimen is kept at the Fujian Agricultural and Forestry University Herbarium (Med-01). Approximately 10 Gb of Pacbio sequence data were extracted from the total sequencing output and input into Organelle PBA (Soorni et al. Citation2017) to assemble the chloroplast genome. Annotation of the chloroplast genome was performed using the Dual Organellar GenoMe Annotator (DOGMA) online tool (Wyman et al. Citation2004) and Geneious ver. 2019.1.1 (Li et al. Citation2019), then manually verified and corrected by comparison with Opisthocentra clidemioides (GenBank accession KX826828).
The complete chloroplast genome sequence of M. dodecandrum (GenBank accession MK836406) was 156,598 bp in length, with a large single-copy (LSC) region of 85,992 bp, a small single-copy (SSC) region of 18,286 bp, separated by two inverted repeat (IR) regions of 26,160 bp each. It was predicted to contain 129 genes, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The overall GC content was 37.2%. While the chloroplast genome sequence of M. dodecandrum from Guangzhou (Zheng et al. Citation2019) was 156,611 bp in length, and the corresponding values in LSC, SSC, and IR regions were 86,015 bp, 17,096 bp, and 26,750 bp, respectively. There may be two reasons for the difference in the length of M. dodecandrum, the different species of reference annotation and the differences between plant populations.
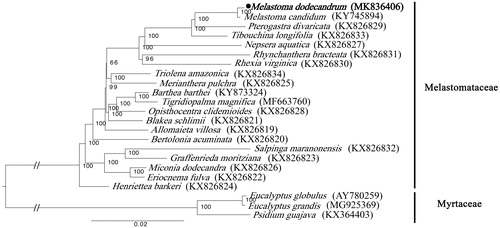
The phylogenetic tree was constructed to demonstrate the relationships between Melastoma and other genera. The chloroplast genome of M. dodecandrum was aligned with other 19 chloroplast genome sequences of Melastomataceae and 3 Myrtaceae species as outgroup, using RAxML (Stamatakis Citation2014) to construct the maximum likelihood tree (). The maximum likelihood tree indicates that M. dodecandrum is phylogenetically close to M. candidum.
Acknowledgements
We are grateful to Dr. Sijin Zeng from South China Agricultural University for his support to this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen J. 2007. Melastomataceae. In: Wu ZY, Raven PH, editors. 2007. Flora of China. Beijing: Science Press; St. Louis: Missouri BotanicalGarden Press. Vol. 13, p. 360–399.
- Editors Commitment. 1984. Flora of China. Vol. 53. Beijing: Science Press.
- Li Y, Li Z, Hu Q, Zhai J, Liu Z, Wu S. 2019. Complete plastid genome of Apostasia shenzhenica (Orchidaceae). Mitochondrial DNA Part B. 4:1388–1389.
- Renner SS, Meyer K. 2001. Melastomeae come full circle: biogeographic reconstruction and molecular clock dating. Evolution. 55:1315–1324.
- Soorni A, Haak D, Zaitlin D, Bombarely A. 2017. Organelle_PBA, a pipeline for assembling chloroplast and mitochondrial genomes from PacBio DNA sequencing data. BMC Genomics. 18:49.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zheng X, Ren C, Huang S, et al. 2019. Structure and features of the complete chloroplast genome of Melastoma dodecandrum. Physiol Mol Biol Plants. https://doi.org/10.1007/s12298-019-00651-x