Abstract
Lonicera oblata, a critically endangered species endemic to North China with about 30 wild individuals, has long been ignored for conservation since its publication because of little attention on its living situation. In this study, we characterized the complete chloroplast (cp) genome of L. oblata. The cp genome was 155,481 bp in length, included a large single-copy (LSC) region of 89,139 bp, a small single-copy (SSC) region of 18,676 bp, and two inverted repeat (IR) regions of 23,833 bp each. The genome contains 130 genes, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic position of L. oblata was also investigated based on cp genome phylogeny of Lonicera representatives. This study is valuable for molecular phylogenetic study and conservation of Lonicera and related taxa.
Lonicera oblata, a critically endangered deciduous shrub belongs to the family Caprifoliaceae, is endemic to northern China which only can be found at stony hillsides with an altitude of about 1000 m. However, this species is in critically endangered condition due to climate change, serious environmental degradation, human disturbance, and low propagation coefficient. At present, just about 30 wild living individuals are found sparsely distributed in Beijing City, Hebei and Shanxi Provinces in China. Although threatened by extinction, the conservation of L. oblata has been ignored for a long time. Genetic diversity is of great importance in conservation plan for endangered plants. In this study, we assembled the complete chloroplast genome of L. oblata and the annotated chloroplast genome sequence has been deposited in the GenBank with an Accession Number MH681655. This study will not only help to investigate population genetic diversity of L. oblata, but also contribute to the molecular phylogeny studies of Lonicera and Caprifoliaceae.
The fresh leaves of L. oblata were collected from Song Mountain, Beijing, China (N 115°43′44″–115°50′22″, E 40°29′9″–40°33′35″). Voucher specimen (collector and collection number: Xian-Yun Mu 3877) is deposited in the herbarium of Beijing Forestry University. Genomic DNA extraction and next-generation sequencing were performed with an Illumina Hiseq platform by Shanghai OE Biotech. Co., Ltd. (Shanghai, China). In total, 16.91 Gb of 150 bp clean reads were generated and used for chloroplast genome assembly through Geneious 11.1.4 software (Kearse et al. Citation2012) with Lonicera japonica (Kang et al. Citation2018) as a reference sequence. The assembled chloroplast genome was then annotated using the Plann (Huang and Cronk Citation2015). Eventually, annotations were verified by Geneious 11.1.4 software.
The complete chloroplast genome of L. oblata was 155,481 bp in length and had a typical quadripartite structure. The genome included a large single-copy (LSC) region of 89,139 bp, a small single-copy (SSC) region of 18,676 bp, and two inverted repeat (IR) regions of 23,833 bp each. Overall, the GC content was 38.4%. A total number of 130 genes in the L. oblata chloroplast genome was identified, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes.
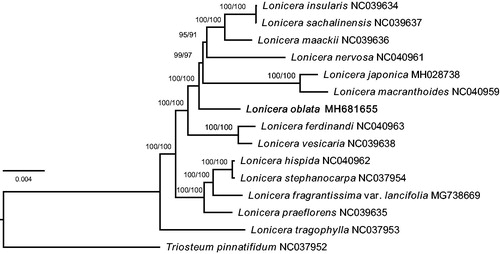
Phylogenetic analysis was conducted using 14 species from Lonicera, and one outgroup specie from Triosteum (). A total of 15 complete chloroplast genomes were aligned using MAFFT (Katoh et al. Citation2017) and adjusted manually. The maximum likelihood (ML) tree was constructed with IQ-TREE software (Kalyaanamoorthy et al. Citation2017; Hoang et al. Citation2018). The result accord with the classification of Subgen. Chamaecerasus and Subgen. Lonicera in traditional classification system, but the phylogenetic relationship intra Subgen. Chamaecerasus needs further investigation.
Disclosure of interest
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35:518–522.
- Huang DI, Cronk QC. 2015. Plann: a command-line application for annotating plastome sequences. Appl Pl Sci. 3:1500026.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods. 14:587–589.
- Kang S, Park H, Koo HJ, Park JY, Lee DY, Kang KB, Han SI, Sung SH, Yang T. 2018. The complete chloroplast genome sequence of Korean Lonicera japonica and intra-species diversity. Mitochondrial DNA Part B. 3:941–942.
- Katoh K, J, Rozewicki KD. Yamada 2017, MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 1–7.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.