Abstract
In this study, we reported the complete mitochondrial genome sequence of a red panda (Ailurus fulgens). The mitogenome (GenBank accession number MK886830) length is 16,517 bp and exhibits the typical structure of mammalian mitochondrial genomes contains 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs, and one control region. The tRNASer (AGY) gene failed to form the typical clover-leaf secondary structure as it lacked the dihydrouridine (DHU) arm. phylogenetically, our sequence cluster together with genus Ailurus, which showed a closer genetic relationship. The mitogenome provides new data to further elucidation and understand the phylogeny of red pandas.
The red panda (Ailurus fulgens) is a carnivorous animal that mainly feeds on bamboo leaves and shoots (Roberts and Gittleman Citation1984). It primarily distributed in China (Tibet, Yunnan, and Sichuan) while the distribution is very narrow due to fragmentation of habitat and Peculiar feeding. Therefore, in view of its population size, the IUCN classified it as Endangered (EN) Species (Glatston et al. Citation2015) and listed as category II species in China. Morphological and molecular studies displayed the classification of red pandas have range relationships to the Ursidae, procyonids and mephitids (Decker and Wozencraft Citation1991; Dragoo and Honeycutt Citation1997; Delisle and Strobeck Citation2005). In this study, we sequenced the complete mitogenome of red panda and constructed Phylogenetic tree, which can be used as a reference for the analysis of the red pandas classification.
The muscle tissue of the red panda was collected from a natural death individual in Hailuogou (N29°36′30″, E102°04′7″) of Gongga Mountains, stored in −80 °C refrigerator of Zoology Laboratory of Sichuan Agricultural University, China. We successfully designed 15 specific primer pairs using the mitochondrial genome of A. ful (AM711897) and Genomic DNAs were extracted by phenol-chloroform protocol (Sambrook et al. Citation1990). We manually edited and assembled the DNA sequences using DNAStar software (Burland Citation1999) and Phylogenetic tree was constructed using Mega 10.0 (Kumar et al. Citation2018).
The newly generate mitogenome sequence (GenBank accession number MK886830) was 16,517 bp and the base composition is A (33%), T (29.54%), C (24.20%), and G (13.27%). Genes arrangement are similar to other mammals that contain 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs, and one control region (Anderson et al. Citation1981). Ten PCGs were started with the typical ATG codon while NADH3 and NADH5 genes initiated with ATT, NADH2 began with ATA. Meanwhile, all PCGs were terminated with typical TAA or TAG, except for COIII, NADH3, NADH4, which ended with incomplete stop codon T––, and CYTB stoped with AGA. In this mitogenome, NADH6 and eight tRNA genes were encoded on the L-strand, whereas the H-strand encoded two rRNAs, 12 PCGs and 14 tRNAs. All tRNAs were folded into the typical clover-leaf secondary structure, except for the tRNASer (AGY) gene as it lacked the dihydrouridine (DHU) arm, which is common phenomenon in other carnivorous (Chen and Zhang Citation2012; Wu et al. Citation2018). 12S rRNA and 16S rRNA were 966 bp and 1575 bp that were separated by the tRNAVal gene. D-loop was located between tRNAPro and tRNAPhe, with a length of 1071 bp.
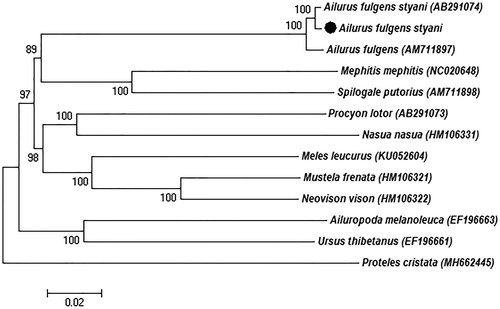
Phylogenetic analysis included the complete mitogenome of three red pandas and other nine species from Caniformia, using Proteles cristata as outgroup (). The neighbour-joining (NJ) analysis exhibited that our sequence cluster together with genus Ailurus and then formed a sister relationship with Mephitidae that consistent with the previous reports (Flynn et al. Citation2005). Furthermore, our study characterized the Ailurus fulgens styani mitochondrial genome sequence, which can be used as a reference for the classification of red pandas and contributes to species conservation.
Figure 1. Neighbour-joining (NJ) phylogenetic tree based on the complete mitochondrial genome of the three red panda sequences and other nine Caniformia species sequences. Proteles cristata was served as outgroup. Numbers at the branches indicated the bootstrapping values with 1000 replications. GenBank accession numbers were given in the parentheses. Filled circle represented a sequence from this study.
Acknowledgements
We thank the Qian Su, Xue Liu, Pu Zhao, Yuhan Wu, Tianrui Xia, Cong Wang, and Kechu Zhang for their assistance in experiments and data analysis.
Disclosure statement
The author reports no conflicts of interest. The author alone is responsible for the content and writing of the paper.
Additional information
Funding
References
- Anderson S, Bankier AT, Barrell BG, Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290:457–465.
- Burland TG. 1999. DNASTAR’S lasergene analysis software. Bioinfromatics methods and protocols, 2nd ed. New Jersey: Humana Press; p. 71–91.
- Chen L, Zhang HH. 2012. The complete mitochondrial genome and phylogenetic analysis of Nyctereutes procyonoides. Acta Ecologica Sinica. 32:232–239.
- Decker DM, Wozencraft WC. 1991. Phylogenetic analysis of recent procvonid genera. Mammal. 72:42–55.
- Delisle I, Strobeck C. 2005. A phylogeny of the Caniformia (order Carnivora) based on 12 complete protein-coding mitochondrial genes. Mol Phylogenet Evol. 37:192–201.
- Dragoo JW, Honeycutt RL. 1997. Systematics of mustelid-like carnivores. J Mammal. 78:426–443.
- Flynn JJ, Finarelli JA, Zehr S, Hsu J, Nedbal MA. 2005. Molecular phylogeny of the Carnivora (Mammalia): assessing the impact of increased sampling on resolving enigmatic relationships. Syst Biol. 54:317–337.
- Glatston A, Wei F, Than Z, Sherpa A. 2015. Ailurus fulgens. The IUCN Red List of Threatened Species. [accessed 2019 April 12]. www.iucnredlist.org.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Roberts MS, Gittleman JL. 1984. Ailurus fulgens. Mammal Species. 222:1–8.
- Sambrook J, Fritsch EF, Maniatis T. 1990. Molecular cloning: a laboratory manual. Anal Biochem. 186:182–183.
- Wu Y, Xu H, Li D, Xie M, Wu J, Wen A, Wang Q, Zhu G, Ni Q, Zhang M, Yao Y. 2018. Complete mitochondrial genome and phylogenetic analysis of a Chinese Eurasian lynx (Lynx lynx). Mitochondrial DNA B. 3:1174–1175.
