Abstract
The first complete mitochondrial genome of Acrossocheilus Labiatus from the Qingshui River were reported in this study with accession number MG878098. The overall nucleotide composition was 31.13% A, 25.10% T, 27.52% C, 16.24% G, respectively. Phylogenetic analysis shows that the A. parallens and A. Hemispinus showed a closest phylogenetic relationship, then clustal with A. Labiatus.
Acrossocheilus is commonly known as monitoring and evaluators of the environmental contaminants by its sensitive to aquatic pollution (Liu et al. Citation2016). In the natural state, Acrossocheilus is widely distributed in the the gravel sediment of rivers and streams. There are more than 10 species belong to Acrossocheilus were reported (Xie et al. Citation2016). Unfortunately, as one of the most important species in Acrossocheilus, Acrossocheilus labiatus is relatively less studied.
For the purpose above, the A. labiatus was obtained from the Qingshui River (N107.87, E26.54) and soaked in ethyl alcohol (95%). The total genomic DNA was extracted from skeletal muscle tissues of the fishes and then stored at −80 °C until use in Guizhou University. Then, 14 pairs of primers were used to amplify contiguous, overlapping segments of the complete mitochondrial genome of A. Labiatus. The complete mitochondrial genome of A. Labiatus has been deposited in the GenBank with accession number MG878098 (16,586 bp in length). The gene arrangement and transcriptional direction were similar to those of the typical teleosts mitogenomes (Chen et al. Citation2012), which included 13 protein-coding genes, 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes and a displacement loop locus (D-loop). As observed in other fish species, the G contents were very low (16.24%) (Yu et al. Citation2016). The others were 31.13%A, 25.10%T, 27.52%C, respectively.
To confirm the phylogenetic placement of A. labiatus in the Acrossocheilus, 11 Acrossocheilus fishes and Cyprinus carpio (out-groups) were chosen. Phylogenetic analyses were constructed by the neighbor-joining (NJ) method performed on both amino acid levels and total mitogenomes (Zou et al. Citation2017) ().
Figure 1. The phylogenetic analyses investigated using N–J analysis indicated evolutionary relationships among 11 Acrossocheilus based on the nucleotide (A) and amino acid (B). Cyprinus carpio (GenBank:KU159761) was used as the outgroup.
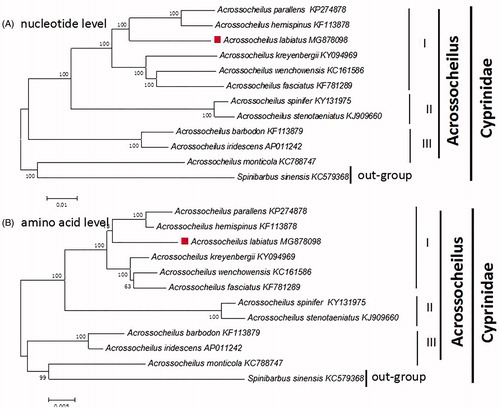
In our study, the phylogenetic placement of A. Labiatus was strongly supported by both amino acid levels and total mitogenomes with the same result (). The 11 Acrossocheilus were divided into three clades (I, II, and III). In group I, there were 6 Acrossocheilus fishes clustal together, while the group II with 2, group III with 2. However, the A. Monticola shows a closest phylogenetic relationship with Cyprinus carpio(out-group) which reveal the Acrossocheilus was not nonmonophyletic. Our work confirmed that the A. Labiatus shows a closest phylogenetic relationship with A. parallens and A. Hemispinus (Xie et al. Citation2016). The A. parallens and A. Hemispinus showed a closest phylogenetic relationship, then clustal with A. Labiatus. The result provided a powerful evidence that the A. Labiatus is a separate species which is in accordance with the previous studies.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen DX, Chu WY, Liu XL, Nong XX, Li YL, Du SJ, Zhang JS. 2012. Phylogenetic studies of three sinipercid fishes (Perciformes: Sinipercidae) based on complete mitochondrial DNA sequences. Mitochondr DNA. 23:70–76.
- Liu GD, Sheng Z, Wang YF, Han YL, Zhou Y, Zhu JQ. 2016. Glutathione peroxidase 1 expression, malondialdehyde levels and histological alterations in the liver of Acrossocheilus fasciatus exposed to cadmium chloride. Gene. 578:210–218.
- Xie XY, Huang GF, Li YT, Zhang YT, Chen SX. 2016. Complete mitochondrial genome of Acrossocheilus parallens (Cypriniformes, Barbinae). Mitochondr DNA A DNA Mapp Seq Anal. 27:3339–3340.
- Yu P, Yang QC, Ding SQ, Zhang J, Li XL, Bi ZH, Wan Q. 2016. The complete sequence of mitochondrial genome of Sinibotia pulchra (Cypriniformes: Cobitidae). Mitochondr DNA A DNA Mapp Seq Anal. 27:4318–4319.
- Zou YC, Xie BW, Qin CJ, Wang MY, Yuan DY, Li R, Wen ZY. 2017. The complete mitochondrial genome of a threatened loach (Sinibotia reevesae) and its phylogeny. Genes Genom. 39:767–778.
