Abstract
We report the complete mitochondrial genome of Sinolapotamon patellifer for the first time, which is found to be 16,547 base pairs in length, and contains 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA), and one non-coding AT-rich region known as the D-loop. In addition, the mitogenome has 17 intergenic regions ranging from 1 to 1512 bp in length. The mitochondrial genome of S. patellifer is the first mitochondrial genome under the genus Sinolapamon, providing DNA data for species identification, enriching the species diversity of Brachyura, and providing a basis for further studies on population genetics and phylogenetics.
The genus Sinolapotamon (Tai Citation1975) (Crustacea: Malacostraca:Decapoda: Brachyura: Potamidae) includes Sinolapotamon patellifer (Wu Citation1934), Sinolapotamon auriculatum (Naruse et al. Citation2010), and Sinolapotamon palmatum (Naruse et al. Citation2010), distributing in mainland China. As a kind of freshwater crab, S. patellifer has no genetic data in NCBI. In order to supplement the vacancy of the gene data of S. patellifer, we report the complete mitochondrial genome for the first time in this paper. The complete mitochondrial genome of S. patellifer will help the identification of species and provide data for further studies on population genetics and phylogenetics.
A male adult specimen of S. patellifer was collected from Chayan Village, Jinbao Township, Yangshuo County, Guilin City, Guangxi Province, China in 2017 (N24°46′49.44″, E110°17′34.08″). The sample has been deposited in the Laboratory Specimen Library of Freshwater Crustacean Decapoda and Paragonimus, School of Basic Medical Sciences, Nanchang University, Nanchang, Jiangxi, PR China and National Parasite Germplasm Resources Specimen Library of China with a catalogue number of JX20174073. The sample was fixed in ethanol and then stored at −20 °C before sequence analyses. Genomic DNA extraction, sequencing, gene annotation, and phylogenetic analyses were performed according to the method described by Plazzi et al. (Citation2013). The Bayesian Inference (BI) method was performed using MrBayes vers. 3.2 (Ronquist et al. Citation2012), with best model GTR + I + G selected using jModelTest vers. 2.1.7. The maximum-likelihood (ML) method was performed using MEGA 6 (Tamura et al. Citation2013).
There are 16,547 base pairs (bp) in the complete mitogenome of S. patellifer (GenBank accession no. MK883709). The genome is rich in AT (76.37% AT content). All genes of the standard metazoan mitogenome are found, including 13 protein-coding genes (PCGs), two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA), and one non-coding AT-rich region known as the D-loop. Among those gens, 23 genes are encoded by the Heavy chain (H chain) and 14 genes are encoded by the Light chain (L chain). The total length of the coding genes of S. patellifer is 14,755 bp, and the total length of the non-coding region is 1729 bp. There are 17 non-coding regions ranging from 1 to 1512 bp in length. The longest non-coding region locates between 12S rRNA and tRNAIle. There are eight overlapping regions, with a total length of 22 bp and the length ranging from 1 to 7 bp in length. The longest gene overlap region is located between ND4 and ND4L.
The complete mitogenome of S. patellifer contains 13 PCGs. Among them, ND1, ND4, ND4L, and ND5 are encoded in the L chain, and the remaining nine encoded protein genes are encoded in the H chain. The initiation codons of ATP6, ND3, and ND6 are ATA, and the initiation codons of the rest PCGs are ATG. The termination codons of all other genes are TAA, except ATP8 (TAG), CYTB (T), and ND5 (T). The average A + T content of the PCGs is 75.18%. ND6 has the highest A + T content of 80.64%, and COX1 has the lowest A + T content of 68.75%.
The lengths of 22 tRNA genes of S. patellifer are between 61 bp (tRNACys) and 72 bp (tRNAVal). Fourteen tRNA genes are encoded on the H chain and the rest is encoded on the L chain. All tRNAs except tRNASer (AGN) exhibited a typical clover structure. The 16S rRNA gene and the 12S rRNA gene are encoded in the L chain and are 1300 bp and 896 bp in length, respectively. The 16S rRNA gene is located between tRNALeu (UUR) and tRNAVa while the 12S rRNA is located between tRNAGln and tRNAIle.
The longest non-coding region of the mitochondrial genome of S. patellifer is located between 12S rRNA and tRNAIle, with a length of 1512 bp and an AT content of 84.79%.
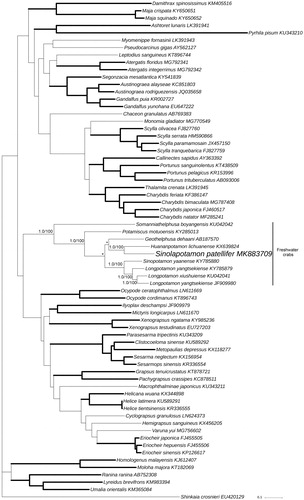
The phylogenetic position of S. patellifer in mitogenome relative to other Brachyuran mitogenomes is determined by applying the BI and ML methods on 13 PCGs (). Our results indicate that as one of the freshwater crabs, S. patellifer is the sister group with Huananpotamon lichuanense. This result is consistent with morphological classification and other molecular analyses (Dai Citation1999; Jia et al. Citation2018).
Figure 1. Bayesian Inference (BI) phylogenetic tree of Sinolapotamon patellifer and other related brachyurans based on 13 PCGs in mitogenomes. Shinkaia crosnieri is served as an outgroup. Numbers on internodes are BI bootstrap proportions and the ML posterior proportions. The same of phylogenetic trees between ML and BI are indicated by bold branches. The differences of freshwater crabs between the ML and BI trees are indicated by ‘*’. The scale bars represent genetic distance.
Acknowledgements
The authors thank Zong-heng Nie (Research lab of Freshwater Crustacean Decapoda & Paragonimus, School of Basic Medical Sciences, Nanchang University, Nanchang, Jiangxi, PR China) for assistance with specimen collection.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Dai AY. 1999. Fauna Sinica, Arthropoda, Crustacea, Malacostraca, Decapoda, Parathelphusidae Potamidae. Beijing: Science Press.
- Jia XN, Xu SX, Bai J, Wang YF, Nie ZH, Zhu CC, Wang Y, Cai YX, Zou JX, Zhou XM. 2018. The complete mitochondrial genome of Somanniathelphusa boyangensis and phylogenetic analysis of Genus Somanniathelphusa (Crustacea: Decapoda: Parathelphusidae). Plos One. 13:e0192601.
- Naruse T, Zhu C, Zhou X. 2010. Two new species of freshwater crabs of the genus Sinolapotamon Tai & Sung, 1975 (Decapoda, Brachyura, Potamidae) from Guangxi Zhuang Autonomous Region, China. Crustaceana. 83:245–256.
- Plazzi F, Ribani A, Passamonti M. 2013. The complete mitochondrial genome of Solemya velum (Mollusca: Bivalvia) and its relationships with Conchifera. BMC Genomics. 14:409.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. English.
- Tai AY. 1975. A preliminary study of the freshwater crabs as intermediate hosts of lung flukes from China. Acta Zool Sinica. 21:169–176.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wu H-W. 1934. Enumeration of the river-crabs (Potamonidae) of China, with descriptions of three new species. Sinensia. 4:338–352.
