Abstract
The complete mitochondrial genome of the ornate threadfin bream, Nemipterus hexodon, was first determined by the pairs-walking sequencing in this study. The circular mtDNA molecule was 17,115 bp in size and the overall nucleotide composition of H-stand was A (29.55%), T (27.36%), G (16.08%), and C (27.01%), with a slight bias towards A + T. The complete mitogenome encoded 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and 2 non-coding regions (an origin of L-strand replication and a control region). The Bayesian tree supported the phylogenetic position of N. hexodon, which provided useful information for phylogenetic relationship in genus Nemipterus.
The ornate threadfin bream, Nemipterus hexodon (Quoy and Gaimard Citation1824), belongs to the family Nemipteridae (Perciformes), which is a warm, small to moderate-sized benthic fish, widely distributed in the Indo-West Pacific Ocean, reaching west to the Andaman Sea, east to Solomon Islands, north to the East China Sea, and south to Australia (Russell Citation1990, p. 37–38). Nemipterus hexodon is one of the artisanal and commercial fisheries in the southeastern coast of China by handline and bottom trawl (Liu et al. Citation2016). It is very common in the coastal fish market in the northern South China Sea and is a popular edible fish species for local people. However, little is known about the fishery resources and fishery biology of N. hexodon and its genetic background has not been reported. Here, we first determined the complete mitogenome sequence of N. hexodon, which was expected to provide insight into the phylogenetic relationship and genetic resources of the species.
One specimen of N. hexodon was collected in May 2010 from Guangdong Leizhou Rare Marine Life National Nature Reserve, Beibu Gulf, the South China Sea (GPS location: 20°39′33″N, 109°44′39″E). It was preserved in 95% ethanol and deposited in Guangdong Ocean University (No. 201003185). Total genomic DNA was extracted from muscle tissue using standard phenol-chloroform method (Sambrook and Russell Citation1989). The complete mitogenome of N. hexodon was obtained by using PCR method, with 15 primer pairs-walking sequencing strategy.
The complete mitochondrial genome of N. hexodon was sequenced to be 17,115 bp in length (GenBank accession number: MK978155). It encoded the canonical 37 genes including 13 protein-coding genes, 22 tRNA genes, and 2 rRNA genes, and 2 non-coding regions (an origin of L-strand replication and a control region). The ND6 and 8 tRNA genes (tRNAGln, Ala, Asn, Cys, Tyr, Ser, Glu, and Pro) were located on L-strand and other genes were transcribed from H-strand. The overall base composition of H-strand was A (29.55%), T (27.36%), G (16.08%), and C (27.01%), with a slight bias towards A + T, which is similar to other threadfin bream (Li et al. Citation2016; Wu et al. Citation2016; Wu and Li Citation2016). Among the 13 protein-coding genes, 12 protein-coding genes started with typical ATG codon, while the COI with GTG codon. Also, 5 protein-coding genes (ND1, COI, ATPase8, ND4L, and ND5) were terminated with TAA codon, ND6 with TAG, and other remaining ones with incomplete stop codon TA– or T–. There were some overlaps among ATPase8 and ATPase6 (10 bp), ND4L, and ND4 (7 bp), ND5 and ND6 (4 bp). The 22 tRNA genes ranged from 67 to 75 bp in size. All tRNA genes could form the typical cloverleaf secondary structures except for tRNASer. The two rRNA genes (12S and 16S rRNA) were 994 bp and 1733 bp in size, respectively and were found between tRNAPhe and tRNALeu, separated by tRNAVal. The origin of L-strand replication (OL) located between tRNAAsn and tRNACys in the WANCY region. The control region (D-loop) was found between tRNAPro and tRNAPhe and was 1357 bp in length, which is shorter than that of Nemipterus bathybius (1603 bp) (Wu et al. Citation2016), but longer than that of Nemipterus japonicus (1260 bp) (Li et al. Citation2016) and Nemipterus virgatus (1260 bp) (Wu and Li Citation2016). Based on the sequence identity analysis of the family Nemipteridae performed in BioEdit version 7.1.9 (Hall Citation1999), the mitogenome sequence of N. hexodon shared 85–86% and 76% identities with that of three Nemipterus species (Li et al. Citation2016; Wu et al. Citation2016; Wu and Li Citation2016) and Scolopsis vosmeri (Wu et al. Citation2017), respectively.
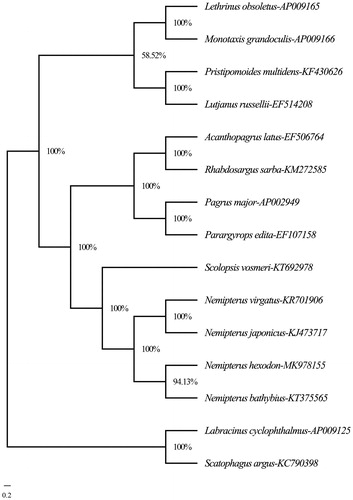
The phylogenetic analysis was constructed by MrBayes version 3.2.7 (Huelsenbeck and Ronquist Citation2001) based on the complete mitogenome sequences of N. hexodon and the other 12 species within four families (Lethrinidae, Lutjanidae, Sparidae, and Nemipteridae) using Labracinus cyclophthalmus (AP009125) and Scatophagus argus (KC790398) as outgroups. The Bayesian tree () showed that N. hexodon first gathered with N. bathybius, then clustered together with N. japonicus and N. virgatus and constituted a monophyly in the family Nemipteridae with S. vosmeri. They formed a sister-group relationship with other three families. Altogether, the results absolutely supported the phylogenetic position of N. hexodon and provided useful information for phylogenetic relationship in genus Nemipterus.
Disclosure statement
The authors report no conflict of interest.
Additional information
Funding
References
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Li YL, Chen JH, Yan BL, Meng XP. 2016. The complete mitochondrial genome of the Japanese threadfin bream, Nemipterus japonicus (Teleostei, Nemipteridae). Mitochondr DNA A. 27:429–430.
- Liu J, Wu RX, Kang B, Ma L. 2016. Fishes of Beibu Gulf. Beijing: Science Press. (Chinese).
- Quoy JRC, Gaimard JP. 1824. Description des Poissons. In: Freycinet, L. De (Ed.), Voyage autour du monde entrepis par ordre du Roi, sous le Ministère et conformément aux instructions de S. Exc. M. Le Vicompte du Bocage, Secrétaire d'état au Départment de la Marine, Exécuté sur les Corvettes de S. M. l'Uranie et al Physicienne, pendant les Années 1817, 1818, 1819 et 1820. Paris: Chez Pillet Aîné, 183–401.
- Russell BC. 1990. Nemipterid fishes of the world. (threadfin breams, whiptail breams, monocle breams, dwarf monocle breams, and coral breams). Family Nemipteridae. An annotated and illustrated catalogue of Nemipterid species known to date. FAO Fisheries Synopsis No. 125, Vol 12. Rome: FAO; p. 37–38.
- Sambrook J, Russell DW. 1989. Molecular cloning: a laboratory manual, 3rd ed. New York: Cold Spring Harbor Laboratory Press.
- Wu ZJ, Li XM. 2016. Complete mitochondrial genome of the Nemipterus virgatus (Perciformes: Nemipteridae). Mitochondr DNA A. 27:3485–3486.
- Wu RX, Liu J, Niu SF, Zheng J. 2017. The complete mitochondrial genome of Scolopsis vosmeri and phylogenetic relationship of genus Scolopsis. Mitochondr DNA A. 28:81–82.
- Wu RX, Niu SF, Qin CX, Liu J. 2016. Complete mitochondrial genome of the yellowbelly threadfin bream, Nemipterus bathybius (Perciformes, Nemipteridae). Mitochondr DNA A. 27:4624–4626.